Ligand-only morgan 1024 experiment¶
In this notebook we will run an experiment using KinoML. In this case we will focus on ligands that have affinity for the kinase EGFR. To run this experiments we will use kinoml.features.ligand.MorganFingerprintFeaturizer, which means that we will only use ligand information to train our model and run our experiments. This notebook is divided into two parts, the first part is about featurizing the dataset and the second part is the actual experiment.
1. Featurize the dataset¶
Any machine learning model will expect tensorial representations of the chemical data. This notebooks provides a workflow to achieve such goal.
kinoml.dataset.DatasetProvider objects need to be available to deal with your collection of raw measurements for protein:ligand systems. These objects are, roughly, a list of kinoml.core.BaseMeasurement, each containing a set of .values and a some extra metadata, like the system objects to be featurized here.
In ligand-based models, protein information is only considered marginally, and most of the action happens at the ligand level. Usually starting with a string representation such as SMILES, or a database identifier such as a PubChem ID, these are promoted to (usually) RDKit objects and then transformed into a tensor of some form (e.g. fingerprints, molecular graph as an adjacency matrix, etc).
Available featurizers can be found under kinoml.features.
[1]:
# If this is the template file (and not a copy) and you are introducing changes,
# update VERSION with the current date (YYYY.MM.DD)
VERSION = "2023.11.06"
[2]:
#fuction to obtain current working path to ouput results
def main():
parameters["HERE"] = str(nbout.parent.resolve())
[3]:
HERE = _dh[-1] #current path
✏ Define hyper parameters¶
Now we will define the parameters of our featurization pipeline. Note that you can adapt these parameters to your data and any experiments you want to run. If you use a different dataset, please make sure it is processed (e.g. no empty values, repeated entries, etc…)
[4]:
# Parameters
DATASET_CLS = "kinoml.datasets.chembl.ChEMBLDatasetProvider"
DATASET_KWARGS = {
"path_or_url": "../data/filtered_dataset_EGFR.csv", #your dataset
"measurement_types": ["pIC50", "pKd", "pKi"], #the type of measurements you are interested in
"uniprot_ids": ["P00533"], #the kinase
"sample": 3000, #number of samples you want to work with, to run the notebook faster you can pick a smaller number
}
PIPELINES = {
"ligand": [
[
"kinoml.features.ligand.MorganFingerprintFeaturizer",
{"nbits": 1024, "radius": 2},
]
]
}
PIPELINES_AGG = "kinoml.features.core.TupleOfArrays"
PIPELINES_AGG_KWARGS = {}
FEATURIZE_KWARGS = {"keep": False}
GROUPS = [
[
"kinoml.datasets.groups.CallableGrouper",
{"function": "lambda measurement: type(measurement).__name__"},
]
]
TRAIN_TEST_VAL_KWARGS = {"idx_train": 0.8, "idx_test": 0.1, "idx_val": 0.1}
#HERE = "."
⚠ If you are adapting this notebook to your own dataset, you should not need to modify anything from here 🤞
Define key paths for data and outputs:
[5]:
from pathlib import Path
HERE = Path(HERE)
for parent in HERE.parents:
if next(parent.glob(".github/"), None):
REPO = parent
break
# Generate paths for this pipeline
featurizer_path = []
for name, branch in PIPELINES.items():
featurizer_path.append(name)
for clsname, kwargs in branch:
clsname = clsname.rsplit(".", 1)[1]
kwargs = [f"{k}={''.join(c for c in str(v) if c.isalnum())}" for k,v in kwargs.items()]
featurizer_path.append("_".join([clsname] + kwargs))
OUT = HERE / "_output" / "__".join(featurizer_path) / DATASET_CLS.rsplit('.', 1)[1]
OUT.mkdir(parents=True, exist_ok=True)
print(f"This notebook: HERE = {HERE}")
print(f"This repo: REPO = {REPO}")
print(f"Outputs in: OUT = {OUT}")
This notebook: HERE = /home/raquellrdc/Desktop/postdoc/kinoml_tech_paper_final/kinoml/examples/experiments/ligand-only-morgan1024-EGFR
This repo: REPO = /home/raquellrdc/Desktop/postdoc/kinoml_tech_paper_final/kinoml
Outputs in: OUT = /home/raquellrdc/Desktop/postdoc/kinoml_tech_paper_final/kinoml/examples/experiments/ligand-only-morgan1024-EGFR/_output/ligand__MorganFingerprintFeaturizer_nbits=1024_radius=2/ChEMBLDatasetProvider
[6]:
# Nasty trick: save all-caps local variables (CONSTANTS working as hyperparametrs) so far in a dict to save it later
_hparams = {key: value for key, value in locals().items() if key.upper() == key and not key.startswith(("_", "OE_"))}
Setup is finished, start working¶
[7]:
from warnings import warn
import os
import sys
from pathlib import Path
from datetime import datetime
import numpy as np
import awkward as ak
from kinoml.utils import seed_everything, import_object
seed_everything();
print("Run started at", datetime.now())
Run started at 2023-11-06 10:51:16.674245
Load raw data¶
This
import_objectfunction allows us to take astrcontaining a Python import path (e.g.kinoml.datasets.chembl.ChEMBLDatasetProvider) and obtain the imported object directly. That’s how we can encode classes in JSON-onlypapermillinputs.See the help message
import_object?for more info. Note that you need an OE license
[8]:
dataset = import_object(DATASET_CLS).from_source(**DATASET_KWARGS)
dataset
/home/raquellrdc/anaconda3/envs/kinoml_env/lib/python3.9/site-packages/pandas/core/computation/expressions.py:21: UserWarning: Pandas requires version '2.8.0' or newer of 'numexpr' (version '2.7.3' currently installed).
from pandas.core.computation.check import NUMEXPR_INSTALLED
[8]:
<ChEMBLDatasetProvider with 3000 measurements (pIC50Measurement=2826, pKdMeasurement=53, pKiMeasurement=121), and 3000 systems (KLIFSKinase=1, Ligand=3000)>
[9]:
#visualise your dataset!
df = dataset.to_dataframe()
df
[9]:
| Systems | n_components | Measurement | MeasurementType | |
|---|---|---|---|---|
| 0 | P00533 & CC(=O)c1ccccc1-n1c(-c2ccccc2)nc2cc(Cl... | 2 | 6.142668 | pIC50Measurement |
| 1 | P00533 & COc1cc(C=O)ccc1-c1cc2c(NCc3ccccc3)ncn... | 2 | 8.221849 | pIC50Measurement |
| 2 | P00533 & O=C(CCN1CCCCC1)Nc1ccc2ncnc(Nc3cccc(Br... | 2 | 9.568636 | pIC50Measurement |
| 3 | P00533 & C=CC(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc... | 2 | 8.397940 | pIC50Measurement |
| 4 | P00533 & N#Cc1cnc(Nc2cccc(Br)c2)c2cc(NC(=O)CCN... | 2 | 5.161151 | pIC50Measurement |
| ... | ... | ... | ... | ... |
| 2995 | P00533 & CN1CCN(c2ccc(NC(=O)c3c(NCCc4cccc(Cl)c... | 2 | 7.366532 | pKiMeasurement |
| 2996 | P00533 & C=CC(=O)Nc1cc(Nc2nccc(-c3cn(C)c4ccccc... | 2 | 8.835647 | pKiMeasurement |
| 2997 | P00533 & Cc1ccc(CNc2nc(N)nc3[nH]c4cc(C)c(O)cc4... | 2 | 5.246417 | pKiMeasurement |
| 2998 | P00533 & COC1CCN(c2nccc(Nc3cc4[nH]c(-c5cn[nH]c... | 2 | 7.119186 | pKiMeasurement |
| 2999 | P00533 & Nc1nc(NCc2ccc(Cl)cc2)c2c(n1)[nH]c1ccc... | 2 | 7.096910 | pKiMeasurement |
3000 rows × 4 columns
Featurize¶
Now, let’s convert the data set into somehting that is ML-readable!
[10]:
# build pipeline
from kinoml.features.core import Pipeline
pipelines = []
for key, pipeline_instructions in PIPELINES.items():
print(f"Building featurizer `{key}` with instructions:")
featurizers = []
for featurizer_import_str, kwargs in pipeline_instructions:
kwargs = kwargs or {} # make sure empty values (None, "") turn into {} so we can do **kwargs below
print(f" Instantiating `{featurizer_import_str}` with options `{kwargs}`")
featurizers.append(import_object(featurizer_import_str)(**kwargs))
pipelines.append(Pipeline(featurizers))
print("Resulting pipelines:", *pipelines)
aggregated_pipeline = import_object(PIPELINES_AGG)(pipelines, **PIPELINES_AGG_KWARGS)
print("Aggregated pipelines:", aggregated_pipeline)
Building featurizer `ligand` with instructions:
Instantiating `kinoml.features.ligand.MorganFingerprintFeaturizer` with options `{'nbits': 1024, 'radius': 2}`
Resulting pipelines: <Pipeline([MorganFingerprintFeaturizer])>
Aggregated pipelines: <TupleOfArrays([Pipeline([MorganFingerprintFeaturizer])])>
[11]:
%%capture --no-display
# to hide warnings
aggregated_pipeline.featurize(dataset.systems, **FEATURIZE_KWARGS);
[11]:
[<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)CC(=O)NCCn1ccc2ncnc(Nc3ccc(Oc4cccc(Cl)c4)c(Cl)c3)c21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C(=O)/C=C/C(=O)NCCCN(C)C)c1cc2c(Nc3cccc(Br)c3)ncnc2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CC[C@H](N2C(=O)N(c3ccccc3)Cc3cnc(Nc4ccc(N5CCN(C)CC5)cc4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)NCCSc1nc(-c2ccccc2)c(-c2ccnc(Nc3ccccc3)c2)[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(-n2cc3c(-c4c[nH]c(-c5ccccc5)c4-c4ccccc4)nnc(O)c3n2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1COc2cc3ncnc(Nc4cccc(F)c4)c3cc2N1CCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(C)n1cnc2cnc(Nc3ccnc(N4CCC(OC)CC4)n3)cc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncc([N+](=O)[O-])n1CCOC(=O)/C=C/c1ccc(-c2ccccc2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CC[C@H](N2C(=O)N(Cc3ccccc3)Cc3cnc(Nc4ccc(N5CCN(C)CC5)cc4OC)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN1CCN(CC/C=C/c2ccc3c(Nc4ccc(Sc5nccn5C)c(Cl)c4)c(C#N)cnc3c2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(C2=NN(c3nc(-c4ccc(Cl)cc4)cs3)C(c3ccc(F)cc3)C2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1CSC(N/N=C/c2cc(Cl)ccc2O)=N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCc1c(C(=O)O)cn2ncnc(Nc3ccc4c(cnn4Cc4ccccc4)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)Nc1ccc2ncnc(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)c1ccc(Nc2nc(NC(=O)C(C)(C)C)nc3[nH]c4cccc(Cl)c4c23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1ccc(C(=O)Nc2nccc(-c3cccnc3)n2)cc1NC(=O)/C=C/CN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@@H](Nc1ncnc2[nH]c(-c3ccc(F)cc3)cc12)c1ccc(F)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Clc1ccc(COC(c2cccs2)c2cc3ccccc3nc2Cl)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNC(=O)c1c([Se][Se]c2[nH]c3ccccc3c2C(=O)NC)[nH]c2ccccc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3cc(OC4CCOCC4)c4c(c23)OCCO4)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#CC(C#N)=C(N)/C(C#N)=C/c1ccc(O)c([N+](=O)[O-])c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#C/C(=C\c1cc(O)c(O)c(CSCc2ccccc2)c1)C(N)=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(Br)cc(-c2nn(C(C)C)c3ncnc(N)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(C)c(O)c(CN(Cc2ccc(O)cc2)C(=S)Nc2ccccc2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NS(=O)(=O)c1ccc(Nc2nc3ncnc(Nc4ccc(N5CCOCC5)c(Cl)c4)c3s2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=C(CN(C)C)C(=O)c1ccc(OS(=O)(=O)c2ccc(C(=O)O)c(O)c2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(-n2c(=O)c(=O)n(C(C)C)c3cnc(Nc4ccc(N5CCN(C)CC5)c(C)c4)nc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)N1N=C(c2ccc(Br)cc2)CC1c1ccc(C(=O)OCCn2c([N+](=O)[O-])cnc2C)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1nc(Nc2cccc(Br)c2)c2cc(Cc3ccc(Cl)cc3Cl)[nH]c2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1ccc2nccc(Nc3cccc(Cl)c3)c2c1)C1CCC2(CC1)OCC1(OO2)C2CC3CC(C2)CC1C3>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(Nc2nc(Nc3ccc(OCCCn4cnnn4)cc3)ncc2[N+](=O)[O-])c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncnc(Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)c1C#Cc1ccc(CN2CCCC2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCCCCCCCCCCCNC(=O)c1cc(NCc2cc(O)ccc2O)ccc1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)NCCSc1nc(-c2ccc(F)cc2)c(-c2ccnc(Nc3ccccc3)c2)[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)n1nc(-c2ccc3cnccc3c2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(NC(=O)c4ccccc4)cc3)ncnc2cc1OCCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc2c(Nc3ccc(F)c(C#N)c3)ncnc2cc1O[C@H]1CCOC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)C/C=C/C(=O)Nc1ccc2c(C#N)cnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CC[C@H](N2C(=O)N(c3cc(OC)ccc3F)Cc3cnc(Nc4ccc(N5CCCCC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CO[C@H]1[C@@H](N(C)C(=O)c2cccc(Cl)c2)C[C@@H]2O[C@@]1(C)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(O)c2c(=O)n(-c3cccc(C(=O)N4CCCCC4)c3)ccc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNC(=O)c1ccc(S(=O)(=O)Oc2ccc(/C=C/[N+](=O)[O-])cc2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCN(CC#CC(=O)Nc1ccc2ncc(C#N)c(Nc3cccc(Br)c3)c2c1)CCC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=S)N1N=C(c2ccc(Br)c(Br)c2)CC1c1ccc([N+](=O)[O-])cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C2=NN(c3nc(-c4ccc(Cl)cc4)cs3)C(c3ccc(Br)cc3)C2)cc1C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1c(Br)cc(/C=C2\C(=O)Nc3ccccc32)cc1C(C)(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)Oc1cccc(Nc2ncnc3cc4oc(=O)n(CCCN5CCOCC5)c4cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(S(=O)(=O)O)cc1.Fc1cccc(COc2ccc(Nc3ncnc4ccc(-c5ccc(CN6CCOCC6)o5)cc34)cc2Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncc(C#N)c(Nc3cc(Cl)ccc3O)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3cccc(C#CCCO)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1[nH]c2ncnc(Nc3cccc(Br)c3)c2c1C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ccc(-c2nn(C3CCCC3)c3ncnc(N)c23)cc1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccn2ncnc(Nc3ccc4[nH]ncc4c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1ccc(Oc2ccnc3[nH]ccc23)c(F)c1)c1nnn(-c2ccc(F)cc2)c1C(F)(F)F>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=S=C(Nc1ccc(Cl)cc1)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2cc3ncc(C#N)c(Nc4ccc(F)c(Cl)c4)c3cc2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N[C@H]1CC[C@@H](n2c(=O)c(-c3ccccc3Cl)c(C)c3cnc(Nc4ccc(N5CCN(C)CC5)c(C)c4)nc32)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(Nc2ncnc3ccc(C(F)(F)F)cc23)cc(OC)c1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2nccc(Nc3ccccc3S(=O)(=O)C(C)C)n2)c(OC)cc1N(C)CCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1NC(=O)CCCCCCC(=O)NO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(NN=C(C#N)C#N)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=[N+]([O-])c1cccc(Oc2cc(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)ncn2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)NC1CCC(n2c(NC(=O)c3cccc(C(F)(F)F)c3)nc3cc(C)ccc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3cc(OC)c(NC(=O)/C=C/CN(C)C4CC4)cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)c1ccc(CCCCCn2c3c(c4cc(OCCc5ccc(O)cc5)ccc42)CC(N)CC3)cc1.Cl.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc2ncnc(N[C@@H](C)c3ccccc3)c2cc1NC(=O)[C@@H]1COC(=O)N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cl.Clc1cccc(Nc2ncnc3ccccc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(N2CCC(N3CCN(S(C)(=O)=O)CC3)CC2)ccc1Nc1nccc(-c2c(-c3cccc(C(=O)Nc4c(F)cccc4F)c3)nc3ccccn23)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1cccc(Oc2ccc(Nc3ncnc4ccn(CCOCCO)c34)cc2Cl)c1)NC1CCCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(Nc2ncnc3cc4c(cc23)N(CCCN2CCCCC2)C(=O)CO4)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/c1ccc(Br)cc1)Nc1ccc2ncnc(Nc3cccc(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(/C=C/C(=O)c2ccc(NC(=O)c3cc4cc5ccccc5nc4s3)cc2)cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncc(C#N)c(Nc3cccc(C(N)=O)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=S=C(NNc1nncc2ccccc12)Nc1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)N1N=C(c2ccc(C)cc2)CC1c1ccc(C(=O)OCCn2c([N+](=O)[O-])cnc2C)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(C2=NN(c3nc(-c4ccc(Cl)cc4)cs3)C(c3ccc(Cl)cc3)C2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)n1nc(-c2cccc3[nH]ccc23)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(Nc2ncc3c(n2)N([C@H]2CCN(C(=O)CN(C)C)C2)C(=O)N(c2ccccc2Cl)C3)ccc1N1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CCCCCCn1cc(-c2ccc3ncnc(Nc4ccc(F)c(Cl)c4)c3c2)nn1)NO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCN([C@H]2CC[C@H](n3nc(-c4ccc(Nc5nc6cccc(Cl)c6o5)c(F)c4)c4c(N)ncnc43)CC2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(N(C)CCN(C)C)c(NC(=O)C=C(C)C)cc1Nc1nc2c(c(-c3cn(C)c4ccc(F)cc34)n1)CS(=O)(=O)CC2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCc1c(C(=O)NC(CO)Cc2ccccc2)[nH]c2ccc(C(F)(F)F)cc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CO/N=C/c1c(N)ncnc1Nc1ccc2[nH]c(Cc3cccc(F)c3)nc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=S)N1N=C(c2ccc(F)cc2)CC1c1ccc2ccccc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(Cl)c1NC(=O)c1cnc(NC(=O)C2CC2)s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)n1nc(-c2ccc3nc(NN)ccc3c2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN1CCN(Cc2ccc(-c3cc4c(N[C@H](C)c5ccccc5)ncnc4[nH]3)cc2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc2ncnc(Nc3cccc(-c4ccco4)c3)c2cc1OCC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)N1CC[C@H](Nc2nc(Nc3ccc(N4CCN(C)CC4)cc3)c3ncn(C(C)C)c3n2)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CCl)Nc1ccc2ncnc(Nc3cccc(I)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Clc1ccc(Nc2cc(Nc3cccc(Cl)n3)ncn2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC1CCC(n2nc(-c3ccc(F)c(O)c3)c3c(N)ncnc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCCCCCCCCCCCCCCCCCNC(=O)COc1cc(O)c2c(=O)cc(-c3ccccc3)oc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(NC3CC3c3ccccc3)c2cc1OCCCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1cccc(Cn2ncc3cc(Nc4ncnn5ccc(COC[C@H]6CNCCO6)c45)ccc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(=O)CCCC(=O)Nc1cc(Nc2cccc(Cl)c2)ncn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)(O)CC(=O)NCCn1ccc2ncnc(Nc3ccc(Oc4cccc(C(F)(F)F)c4)c(Cl)c3)c21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=C(C)C(=O)Cc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc2cc(Nc3ccnc4cc(-c5ccc(CNCCO)cc5)sc34)ccc2[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc(Nc2ccc3c(cnn3Cc3cccc(F)c3)c2)c1/C=N/NCC(F)(F)F>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(-c2c[nH]c3nccc(-c4[nH]c(CCCO)nc4C(F)(F)F)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#Cc1cnc2ccc(-c3ccc(CN)cc3)cc2c1N[C@@H]1C[C@H]1c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CCC[C@@H](N2C(=O)N(Cc3ccccc3)Cc3cnc(Nc4ccc(N5CCN(C)CC5)cc4OC)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(C#N)cc3)ncnc2cc1OCCCN1CCC(c2ccccc2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1CN1CCC[C@@H]1C(N)=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ccccc1NC(=O)/C=C/c1ccc(-c2ccc3ncnc(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)c3c2)o1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(C(/C=C/c2ccccc2Cl)=N\NC(N)=S)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNC(=O)c1c(SSc2[nH]c3ccccc3c2C(=O)NC)[nH]c2ccccc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncc([N+](=O)[O-])n1CCOC(=O)/C=C/c1ccccc1F>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#CC(C#N)=C1CCc2c1ccc(O)c2O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(/N=N/c1ccc2ncnc(Nc3cccc(Cl)c3)c2c1)C(=O)Oc1ccc([N+](=O)[O-])cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1ccc(Oc2nc(Nc3ccc(N4CCN(C)CC4)cc3OC)ncc2Cl)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2nccc(Nc3ccccc3-n3nccn3)n2)c(OC)cc1N1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Oc1ccc(CNc2ccc3ncnc(Nc4cccc(Cl)c4)c3c2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=c1ccc(OCc2nnc(Cc3nc4ccccc4[nH]3)o2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc2c1c(-c1cccc(O)c1)cn2C1CCNC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccn2ncnc(Nc3ccc4c(cnn4Cc4ccccn4)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=c1ccc(Cn2ncc3cc(Nc4ncnn5ccc(COC[C@@H]6CNCCO6)c45)ccc32)nc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cccc(-c2cn(C3CCN(CCO)CC3)c3ncnc(N)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)CCN(C)c1c(Br)cccc1Nc1ncnc2ccncc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OCCN(CCO)CCNc1cc2c(Nc3cccc(Br)c3)ncnc2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1nn2c(=O)cc(-c3ccccc3)[nH]c2c1/N=N/c1ccc2c(c1)OCO2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCOC(=O)Nc1ccc(Nc2ncnc3cc(OC)c(OC)cc23)cc1C.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCc1c(C(N)=O)cn2ncnc(Nc3ccc4c(cnn4Cc4ccccc4)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1cccc2c(Nc3cccc(Br)c3)ncnc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(N(C)C(=O)c2ccccc2)cc1Nc1nccc(-c2cccnc2)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2nccc(-c3cn(C)c4cccnc34)n2)c(OC)cc1N(C)CCNC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc(Nc2ccc3c(cnn3Cc3cccc(F)c3)c2)c1/C=N/N1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#CC(C#N)=Cc1cc[nH]n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(=O)c1ccc(NC(=O)CSc2nnc(CNc3ccc(OC)cc3)n2-c2ccccc2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@@H](Nc1ncnc2sc(-c3ccc(F)c(C=O)c3)cc12)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OB(O)c1ccc2ncnc(Nc3cccc(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)N(CC#CC(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cn1)C(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COCC(=O)Nc1ccc2ncnc(Nc3cc(Cl)c(Cl)cc3F)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Brc1cccc(Nc2ncnc3sc4c(c23)CCCC4)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2ncnc(-c3cn(C)c4ccccc34)c2OC)c(OC)cc1N(C)CCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COCC(C)Oc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2c2c1OCCO2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cccc2ncnc(Nc3ccc(OCc4ccccn4)c(Cl)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC[C@@H](Nc1ncnc2oc(-c3ccccc3)cc12)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Cc1ccc(Br)cc1)NS(=O)(=O)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ncc(-c2nn(C(C)C)c3ncnc(N)c23)c(OC)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1COc2cc3ncnc(Nc4ccccc4)c3cc2N1CCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(NC(=O)[C@@H](c4ccccc4)N4Cc5ccccc5C4=O)cc3F)ncnc2cc1OCC1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1OCc1cn(Cc2ccccc2F)nn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(C(=O)NCCN(C)C)cc1-c1cc2c(N[C@H](C)c3ccccc3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(Oc2ccc(Nc3ncnc4cc(C#C[C@@H]5C[C@@H](OC(=O)N6CCOCC6)CN5)sc34)cc2Cl)no1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1ccc(COC(c2cccs2)c2cc3ccccc3nc2Cl)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C(=O)N(CCCl)N=O)c1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC1(C)OOC2(CCC(C(=O)Nc3ccc4ncnc(Nc5cccc(Cl)c5)c4c3)CC2)OO1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3ccc(-c4ccc(CO)o4)cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC[C@H]1Oc2cc3ncnc(Nc4cccc(Cl)c4)c3cc2O[C@@H]1COC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(=O)N1CCN(CCC(=O)Nc2cccc(Oc3nc(Nc4ccc(N5CCN(C)CC5)cc4OC)ncc3Cl)c2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)CCCOC(=O)/C=C/C(=O)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=[O-][S+]1CCN(Cc2ccc(-c3cc4c(Nc5ccc(OCc6cccc(F)c6)cc5)ncnc4cn3)o2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN(CC)C/C=C/C(=O)N1CCc2c(sc3ncnc(N[C@H](CO)c4ccccc4)c23)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(Nc2ncc3cc(-c4c(Cl)cccc4Cl)c(=O)n(C)c3n2)cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN(C/C=C/C(=O)Nc1cccc(-c2c(-c3ccccc3)oc3ncnc(N[C@H](CO)c4ccccc4)c23)c1)CCO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NP(=O)(OCCCOc1ccc2ncnc(Nc3cccc(Br)c3)c2c1)N(CCCl)CCCl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(c1ccccc1)c1cccc(NC(=O)c2cc3ccccc3[nH]2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(Cl)cc3F)ncnc2cc1OC[C@@H]1CCCN(C)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@@H](Nc1[nH]cnc2nc(-c3ccccc3)cc1-2)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(NS(=O)(=O)c2ccc(Nc3nncc4ccccc34)cc2)no1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(N2CCN(C)CC2)ccc1Nc1ncc(Cl)c(Oc2cccc(NC(=O)CCN3CCCC3)c2)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(NCc3ccccc3)ncnc2cc1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(=O)C1=C(N)N(c2cccnc2)C2=C(C(=O)CC(C)(C)C2)C1c1c(C)nn(-c2ccc(Cl)cc2)c1Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#CC(C#N)=C1CCc2cc(O)c(O)cc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc2ncc(C#N)c(Nc3cccc(Br)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(N2CCN(C)CC2)ccc1Nc1ncc(Cl)c(Oc2ccccc2NC(=O)COCc2ccccc2)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccccc1-c1cn(-c2ccccc2)c2ncnc(N)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(F)cc(-n2c(=O)cc(C)c3cnc(Nc4ccc(N5CCN(C)CC5)cc4OC)nc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1nccc(-c2c(-c3ccc(F)cc3)ncn2C2CCCCC2)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cl.c1ccc(Nc2[nH]cnc3c4ccccc4nc2-3)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CC[C@H](N2C(=O)N(c3cc(OC)ccc3F)Cc3cnc(Nc4ccc(N5CCN(C)CC5)c(Cl)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=S=C(NCc1ccccc1)Nc1ccc2ncnc(Nc3cccc(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Brc1cccc(Nc2ncnc3cc(NCCCn4ccnc4)ncc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(Nc2nc(Nc3ccc(CN4CCOCC4)cc3)ncc2F)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)c2cc1OCCCN1CCNCCNCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncnc(Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)c1C#CCOc1ccc(CNC(C)C)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)NCCNCc1ccc(-c2cc3nccc(Nc4ccc5[nH]ccc5c4)c3s2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCn1nc(-c2ccc(OC)c(O)c2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C(=O)Nc2ccc(CN3CCN(C)CC3)c(C(F)(F)F)c2)cc1NC(=O)c1cnc(NC2CC2)nc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(NC(=O)Nc4ccc(Cl)cc4)c(Cl)c3)ncnc2cc1OCCCN1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cl.c1ccc(-c2cccc(Nc3ncnc4cc5c(cc34)OCCCO5)c2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(NC(=O)Nc2ccnc(Nc3ccc(N4CCN(C)CC4)cc3OC)n2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc2c(Cc3ccc(F)c(Cl)c3)ncnc2cc1OCCOCCCCO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(=O)C(=CNc1ccc2ncnc(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)c2c1)C(=O)OCC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCN(Cc2ccc(C(=O)Nc3ccc(-c4cncc(C#N)c4Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)cc3)cc2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1ccc(CN2CCCCC2)cc1)Nc1ccc2ncnc(Nc3cccc(C(F)(F)F)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CC[C@H](N2C(=O)N(c3ccccc3)Cc3cnc(Nc4ccc(CCN5CCN(C)CC5)cc4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@H](COc1cccc2ncnc(Nc3ccc(OCc4ccccn4)c(Cl)c3)c12)N(C)C(=O)CO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1ccc2ncnc(Cc3ccc(F)c(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ccc(C#Cc2cncnc2Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(Nc2[nH]nc3ncnc(Nc4cccc(Cl)c4)c23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCc1c(C(=O)OCC)cn2ncnc(Nc3ccc4c(cnn4Cc4ccccc4)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCC(c2c(O)cc(O)c3c2O/C(=C\c2ccccc2Cl)C3=O)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CS(=O)(=O)Nc1cccc(-c2c(-c3ccccc3)oc3ncnc(N[C@H](CO)c4ccccc4)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCOC(=O)Nc1ccc(Nc2ncnc3cc(OC)c(OC)cc23)cc1Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)C/C=C/C(=O)Nc1cc2c(Nc3cccc(Br)c3)c(C#N)cnc2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NS(=O)(=O)c1ccc(Nc2nc3ncnc(Nc4c(F)cccc4F)c3s2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Cc1cc(F)c(F)cc1F)Nc1cccc(Oc2cc(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)ncn2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)N1N=C(c2ccc(Br)cc2)CC1c1ccc(C(=O)OC(C)Cn2c([N+](=O)[O-])cnc2C)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(/C=C(\C#N)C(N)=O)cc(CSCc2ccc(Cl)cc2)c1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=O)c1cnc2ccc(C(=O)/C=C/c3ccc(Cl)cc3)cc2c1Nc1ccc(Br)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNC(CS(C)(=O)=O)c1ccc(-c2ccc3ncnc(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)c3c2)o1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=O)Nc1ccc(C(=O)Nc2ccc3ncnc(Nc4ccc(F)c(Cl)c4)c3c2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(O)C(/N=C1/C=C(O)/C(=N\C(C(=O)O)C(C)O)C=C1O)C(=O)O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(NC(=O)Nc2ccnc(Nc3ccc(N4CCC(N5CCOCC5)CC4)cc3OC)n2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(C2=NN(C3=NC(=O)CS3)C(c3ccccc3)C2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2ncc(Cl)c(-c3cnn4ccccc34)n2)c(OC)cc1N1CCN(C(=O)N2CCNCC2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CCN1CCCCC1)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/CN1CCCC1)Nc1cc2c(Nc3ccc(Cc4ccccn4)cc3)ncnc2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)C(CO)Nc1nc(Nc2ccc(N3CCNCC3)cc2)c2ncn(C(C)C)c2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(-c2cn(C3CCCC3)c3ncnc(N)c23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)CCNCc1ccc(-c2cc3c(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)ncnc3s2)o1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)CCNCc1nc(-c2ccc3ncnc(Nc4ccc(F)c(Cl)c4)c3c2)cs1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CO/N=C/c1c(N)ncnc1Nc1ccc2c(c1)CCN2Cc1cccc(F)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/CN1CCC(F)(F)C1)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cl.NC1CCc2c(c3cc(OCCc4ccc(O)cc4)ccc3n2CCCCCc2ccccc2)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@@H](Nc1ncnc2sc(-c3ccc(C(=O)O)cc3)cc12)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2nccc(-c3cn(C)c4ccc(F)cc34)n2)c(OC)cc1N(C)CC[N+](C)(C)[O-]>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1nc(Nc2cccc(Br)c2)c2ccn(Cc3ccc(Cl)cc3)c2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(-n2c(=O)cc(C)c3cnc(Nc4ccc(N5CCN(C)CC5)cc4OCC)nc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(C=O)ccc1-c1cc2c(NCc3ccccc3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(C=O)ccc1-c1cc2c(Nc3ccccc3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(NS(=O)(=O)c1ccc(F)cc1)c1cncc(Br)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCC(=O)NO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=c1csc(-c2cc3ncnc(Nc4ccc5[nH]ccc5c4)c3s2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(Nc2ccnc(Nc3ccc(N4CCN(C)CC4)cc3)n2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCNC(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC1CCN(Cc2ccn3ncnc(Nc4ccc5c(cnn5Cc5ccccn5)c4)c23)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/c1ccccc1)Nc1cc2c(Nc3cccc(Br)c3)ncnc2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN(CC)[C@@](C)(C#Cc1ncnc2cc(OC)c(OC)cc12)CC1CCCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(Nc2ncnc3cc(OC)c(OC)cc23)cc(OC)c1.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCc1ccc(C(=O)/C=C\C(=O)O[C@H](CC=C(C)C)C2=CC(=O)c3c(O)ccc(O)c3C2=O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC1(CO)CCN(c2nccc(Nc3cc4c(cn3)nc(C)n4C(C)C)n2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccccc1Nc1cc[nH]c(=O)c1-c1nc2c(C(C)C)ccc(C)c2[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3cc(OCCOC(=O)CC4=C(C)/C(=C\c5ccc([S+](C)[O-])cc5)c5ccc(F)cc54)c(OCCOC)cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CC[C@@H](Nc2nc(Nc3cnn(C)c3)nc3[nH]cc(Cl)c23)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncnc(Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)c1C#CCCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3ccc(NC(C)=O)cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=C(CN1CCNCC1)C(=O)c1ccc(OCc2ccccc2)cc1.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)Nc1ccc(-c2c(-c3ccccc3)oc3ncnc(N[C@H](CO)c4ccccc4)c23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)/N=N/c1ccc2ncnc(Nc3cccc(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc2c(Nc3cccc(Br)c3)ncnc2cc1SCCCN(CC)CC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NS(=O)(=O)c1ccc(Nc2nc3ncnc(Nc4ccccc4)c3s2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccccc1Nc1nc2cc(F)c(N(C)C(=O)/C=C/CN(C)C)cc2n2cncc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(NC(=O)c2ccc(N(CCCl)CCCl)cc2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CC[C@H](N2C(=O)N(c3cc(OC)ccc3F)Cc3cnc(Nc4ccc(N5CCOCC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1ccc2c(c1)CNc1c(Nc3cccc(Br)c3)ncnc1O2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(NC(=O)c4ccccc4)cc3)ncnc2cc1OCCN1CCCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCN1CC2(CSC2)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CO[C@H]1[C@@H](N(C)C(=O)OC(C)(C)C)C[C@@H]2O[C@@]1(C)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC1CCN(C/C=C/C(=O)N2CCOc3cc4ncnc(Nc5ccc(F)c(Cl)c5)c4cc32)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(CCO)c1cc2ncnc(Nc3cccc(Br)c3)c2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc2occ(-c3ccc(NC(=O)Nc4cc(C(F)(F)F)ccc4F)cc3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CC[C@@H](Nc2nc(Nc3ccccc3)nc3cnc(Nc4ccc(N5CCN(C)CC5)cn4)cc23)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(F)cc(Nc2cc(NC(=O)Nc3c(Cl)cccc3Cl)ncn2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Cc1ccc(Cl)cc1)NS(=O)(=O)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)CCNCc1ccc(-c2ccc3ncnc(Nc4ccc(OCc5ccccc5)cc4)c3c2)o1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CO[C@H]1[C@@H](N(C)C(C)=O)C[C@@H]2O[C@@]1(C)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1nccc(C#Cc2cncnc2Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Oc3ccc(NS(=O)(=O)c4ccc(-c5cnn(C)c5)cc4)cc3F)ccnc2cc1OCCCN1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CC[C@H](N2C(=O)N(c3ccccc3Cl)Cc3cnc(Nc4ccc(N5CCN(C)CC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1cccc(COc2ccc(Nc3ncnc4cc(-c5ccc[nH]5)sc34)cc2Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2nccc(NC(=O)c3ccc(OC)c(OC)c3)n2)c(OC)cc1N1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(NCc3cccc(C)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(Br)c(/C=C2\CC(C)C/C(=C\c3nc(C)c(C)nc3C)C2=O)cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1CSC(N/N=C/c2ccc(Br)cc2)=N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc(Nc2ccc(OCc3ccccc3)c(Cl)c2)c1C(=O)Nc1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(Nc2cc(Nc3cccc(Br)c3)ncn2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC/C=C\C(=O)Nc1cc(Nc2nc(C)cc(-c3cn(C)c4ccccc34)n2)c(OC)cc1N(C)CCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)Nc1cccc(Nc2cc(-c3[nH]c(SC)nc3-c3ccc(F)cc3)ccn2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncc(C=O)c(Nc3cccc(Br)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOCc1ccc2c(c1)nc(NC(=O)c1cccc(C(F)(F)F)c1)n2[C@H]1CC[C@H](O)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CCC[C@@H](n2c(=O)c(-c3cc(F)ccc3Cl)c(C)c3cnc(Nc4ccc(N5CCN(C)CC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CCN1CCOCC1)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N(c1ccc(F)c(C)c1)c1ncnc2cc(OC)c(OCCCN3CCOCC3)cc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(=O)CCCn1c(=O)oc2cc3ncnc(Nc4ccc(OC)cc4)c3cc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(/C=N/NC2=NC(=O)CS2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3cc4c(cc23)N(C(=O)/C=C/CN2CCCC2)CCO4)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(Nc2ncnn3ccc(CN4CCC(N)CC4)c23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc2c1c(-c1ccc(Br)c(O)c1)nn2C1CCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1OCc1cn(Cc2cccc(F)c2)nn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CCC[C@@H](Oc2nc(Nc3ccc(N4CCN(C(C)=O)CC4)cc3)nc3[nH]cc(-c4cncc(NC)n4)c23)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NP(=O)(OCCCCOc1ccc2ncnc(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)c2c1)N(CCCl)CCCl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Oc1cccc(Nc2[nH]nc3ncnc(Nc4cccc(Cl)c4)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)N1N=C(c2ccc(C)c(C)c2)CC1c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNC(=O)c1c([Se][Se]c2c(C(=O)NC)c3ccccc3n2C)n(C)c2ccccc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1nc(N)c2c(Nc3cccc(Cl)c3)[nH]nc2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CC[C@H](N2C(=O)N(c3cccc(OC)c3Cl)Cc3cnc(Nc4ccc(N5CCN(C)CC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnn3ccc(CN4CCC(N)CC4)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1cccc(Nc2cc(Nc3cccc(C(F)(F)F)c3)ncn2)c1)C1CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1cnn(-c2ccccc2)c1)c1cccc(NS(=O)(=O)c2ccccc2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1O[C@H]1CCOC1)N1CCC(CCO)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OC[C@@H](Nc1ncnc2sc(-c3ccccc3)cc12)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#Cc1cnc(Nc2cccc(Br)c2)c2cc(NC(=O)CCN3CCOCC3)ccc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=[N+]([O-])/C=C/c1ccc(O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCOC(=O)Nc1ccc(Nc2ncnc3cc(OC)c(OC)cc23)cc1C.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#C/C(=C/c1cc(Cc2ccccc2)c(O)c(Cc2ccccc2)c1)C(N)=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)C/C=C(\F)C(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1O[C@H]1CCOC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C/C(=N\Nc1ncc(Cl)c(Nc2cccc(NC(=O)/C=C/CN(C)C)c2)n1)c1ccc(F)cc1F>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(-c2nn(C(C)C)c3ncnc(N)c23)ccc1NC(C)CO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(NS(=O)(=O)c1ccc(Br)cc1)c1ccc(Cl)nc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NCCCCOCc1ccn2ncnc(Nc3ccc4c(cnn4Cc4cccc(F)c4)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CC[C@H](N2C(=O)N(c3cc(OC)ccc3Cl)Cc3cnc(Nc4ccc(N5CCN(C6COC6)CC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=c1nc(Nc2ccc3[nH]ccc3c2)c2sccc2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2ncc(Cl)c(-c3cnn4ccccc34)n2)c(OC)cc1N1CCN(C(=O)[C@H](C)O)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)(CO)NCc1ccc(-c2cc3ncnc(Nc4ccc5[nH]ccc5c4)c3s2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ccc2c(Nc3cccc(C(F)(F)F)c3)ncnc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3c(F)cc(Cl)cc3F)ncnc2cc1OCC1CCNCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CCC[C@@H](n2nc(-c3cccc(F)c3)c3c(N)ncnc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc2c1c(-c1cccc(O)c1)cn2C1CCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(NC(=O)Nc2nc(Nc3ccc(N4CCC(N5CCOCC5)CC4)cc3OC)ncc2Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(OC)c(Cc2cc3c(Nc4ccc(Cl)cc4F)nc(N)nc3[nH]2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)CCC(=O)c1ccc(OCc2ccccc2)c(O)c1.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(-n2cc(CN3CCNCC3)nn2)ccc1Nc1ncc(Cl)c(Nc2ccccc2S(=O)(=O)C(C)C)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(-n2c(=O)cc(C)c3cnc(Nc4ccc(N5CCN(C)CC5)cc4OC)nc32)cc(C(F)(F)F)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc2ncnc(Nc3cccc(Cl)c3)c2cc1OCC.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN(CC)CCNC(=O)c1cc2cnnc(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)c2[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(Nc2nc(Nc3ccc(OCCN4CCOCC4)cc3)ncc2Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNc1ncc2ncnc(Nc3cccc(C)c3)c2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncc([N+](=O)[O-])n1CC(=O)NS(=O)(=O)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1cccc(COc2ccc(Nc3ncnc4sc(-c5ccco5)cc34)cc2Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)N1N=C(c2ccc(Cl)c(Cl)c2)CC1c1ccccc1Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1NC(=O)CO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(Br)c(/C=C2\CC(C)CC(=C\c3nc(C)c(C)nc3C)/C2=N/O)cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Clc1ccc(Nc2ncc3c(n2)-c2ccc(Cl)cc2SC3)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OCCn1ccc2ncnc(Nc3ccc(Oc4cccc5sncc45)c(Cl)c3)c21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ccc2c(c1)CNc1c(Nc3cccc(Br)c3)ncnc1O2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc2ncc(C#N)c(Nc3cccc(Br)c3)c2cc1OCC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(-n2cc(CN3C[C@H](C)N[C@H](C)C3)nn2)ccc1Nc1ncc(Cl)c(Nc2ccccc2S(=O)(=O)C(C)C)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(CC(O)CO)c1c(Br)cccc1Nc1ncnc2ccncc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Clc1cccc(Nc2[nH]nc3ncnc(Nc4cccc(Br)c4)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(C)c(Br)c3)ncnc2cc1OCCOCCNC(=O)Cn1cnc([N+](=O)[O-])n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3cccc(Cl)c3F)c2cc1CN1CCC1C(N)=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc2nccc(Nc3cccc(Br)c3)c2cc1NC(=O)/C=C/CN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CCl)OCCn1c(=O)oc2cc3ncnc(Nc4ccc(Oc5cccc(C(F)(F)F)c5)c(Cl)c4)c3cc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(Oc2nc(Nc3ccc(N4CCN(C(=O)CC)CC4)cc3OC)ncc2SC)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc(Nc2ccc(OCc3ccccc3)c(Cl)c2)c1C(=O)NNC(=O)CCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC(=O)c1ccc(OC)c(-c2cc3c(N[C@H](C)c4ccccc4)ncnc3s2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CO[C@H]1[C@@H](N(C)C(=O)c2cccc([N+](=O)[O-])c2)C[C@@H]2O[C@@]1(C)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCNCC#CC(=O)Nc1ccc2ncc(C#N)c(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)CCNCCCCOc1ccc2ncnc(Nc3ccc(F)c(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Clc1cccc(Nc2ncnc3nc(Nc4ccccc4)sc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@@H](Nc1ncnc2oc(-c3ccc(NCCN4CCCCC4)cc3)cc12)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc(O)c2c(=O)cc(-c3ccccc3)oc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Oc1ccc(C2CC(c3ccc(Br)cc3)=NN2c2nc(-c3ccc(Cl)cc3)cs2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccccc1NC(=O)/C=C/c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(NC(=O)Nc2ccnc(Nc3ccc(C4CCN(C)CC4)cc3)n2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(-n2cc(NC(=O)c3cccc(NS(=O)(=O)c4ccccc4)c3)cn2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCN1CCCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncnc(Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)c1/C=C/c1ccc(CNCCS(C)(=O)=O)o1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#Cc1cnc(Nc2cccc(Br)c2)c2cc(NC(=O)c3ccco3)ccc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/CN1CCCCC1)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(-c4nc5ccccc5s4)cc3)ncnc2cc1OCCCN1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=c1ccc(CNc2ncnc3oc(-c4ccccc4)cc23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(=O)n2nc(SCc3cccc([N+](=O)[O-])c3)nc2[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1ccc(Nc2ncnc3cc4c(cc23)OCCOCCOCCO4)cc1Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(C2=C(c3c[nH]c4ccccc34)CNC2=O)cc(OC)c1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC[C@@H](Nc1ncnc2sc3c(c12)CCN(C(=O)/C=C/CN(C)C)C3)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cn1nc(-c2ccc3ccccc3c2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1NC(=O)c2cc(Nc3ccc(O)cc3)c(Nc3ccc(O)cc3)cc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=O)CN1CCC(n2cc(-c3cccc(O)c3)c3c(N)ncnc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(CC(=O)NS(=O)(=O)c2ccc(F)cc2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Brc1cccc(Nc2ncnc3c2NCc2ccccc2O3)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(-c4nc5ccc(Cl)cc5s4)cc3)ncnc2cc1OCCCN1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCC(O)(c2nc(-c3ccc(F)cc3)c(-c3ccncc3)o2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2cc(-c3cn4c5c(cccc35)CCC4)nc(OC)n2)c(OC)cc1N(C)CCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@@H](O)c1nc2cnc(Nc3ccnc(N4CC[C@H](O)[C@H](F)C4)n3)cc2n1[C@H](C)C(F)(F)F>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OC[C@@H](Nc1ncnc2oc(-c3ccccc3)c(-c3ccccc3)c12)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1cccc(Cn2ncc3cc(Nc4ncnn5ccc(CNC6CCNCC6)c45)ccc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Brc1cccc(Oc2ncnc3ccccc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/c1ccccc1)Nc1cc(F)cc(F)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(Nc2cc(N3CCN(C)CC3)nc(Oc3ccc(N)cc3)n2)n[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C/N=N/Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@H](NC(=O)c1ccc(S(=O)(=O)Oc2ccc(/C=C/[N+](=O)[O-])cc2)cc1)C(=O)N[C@@H](C)C(=O)NCCN.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(N2CCOCC2)cc2[nH]c(-c3c(NC[C@@H](O)c4cccc(Cl)c4)cc[nH]c3=O)nc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)n1nc(-c2ccc3occc(=O)c3c2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CC[C@H](N2C(=O)N(c3ccccc3)Cc3cnc(Nc4ccc(N5CCN(C)CC5)c(F)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)OCOC(=O)N(C)/N=N/c1ccc2ncnc(Nc3cccc(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(C2=NN(C(N)=S)C(c3ccc4ccccc4c3)C2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2ncc(Cl)c(Nc3ccccc3P(C)(C)=O)n2)c(OC)cc1N(C)CCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(N2NC(=O)/C(=C/c3ccc(-c4ccccc4F)o3)C2=O)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=S)N/N=C(/C=C/c1ccccc1Br)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)CCNc1nc(-c2ccc3ncnc(Nc4ccc(OCc5ccccc5)c(Cl)c4)c3c2)cs1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc2ncnc(Nc3cccc(Br)c3)c2cc1OCC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=COC(=O)N(CCN(C)C)/N=N/c1ccc2ncnc(Nc3cccc(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=S)N1N=C(c2ccc(Cl)c(Cl)c2)CC1c1cccc2ccccc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cccc(-c2nn(C(C)C)c3ncnc(N)c23)c1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(OC)c2c(=O)n(-c3cccc(C(=O)N4CCCCC4)c3)ccc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/c1ccccc1)Nc1cccc(Oc2cc(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)ncn2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)Nc1ccc2c(c1)CNc1c(Nc3cccc(Br)c3)ncnc1O2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1ccc(-n2c(=O)cnc3cnc(Nc4ccc(N5CCN(C)CC5)cc4OC)nc32)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=S(=O)(O)CCNc1cc2ncnc(Nc3cccc(Br)c3)c2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1nc2c(c(Nc3cccc(Br)c3)n1)-c1ccccc1C2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1nc(Nc2cccc(Br)c2)c2cc(Cc3ccc4ccccc4c3)[nH]c2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1Cc2cccc(Oc3ccc(Nc4ncnc5ccn(CCO)c45)cc3Cl)c2N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#C/C(=C\c1ccc(O)c(O)c1)C(=O)NCCCCCCCCCCNC(=O)/C(C#N)=C/c1ccc(O)c(O)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC(=O)CN1CCC(n2cc(-c3cccc(O)c3)c3c(N)ncnc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)c1ccc(Nc2ncnc3ccncc23)c(N)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COCCn1c(=O)oc2cc3ncnc(Nc4ccc(F)c(F)c4)c3cc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)c1cc2ncnc(Nc3cccc(Br)c3)c2cc1[N+](=O)[O-]>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1nc2cnc(Nc3ccnc(N4CCC(S(C)(=O)=O)CC4)n3)cc2n1C(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/CN1CCOCC1)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CC[C@H](N2C(=O)N(c3cc(OC)ccc3F)Cc3cnc(Nc4ccc(N5CCN(C(C)=O)CC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(CCCl)C(=O)Nc1ccc2ncnc(Nc3cccc(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CCC[C@@H](n2nc(-c3cccc(C(F)(F)F)c3)c3c(N)ncnc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COCCNCCOc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3cc(OCCOC(=O)CC4=C(C)/C(=C/c5ccc([S+](C)[O-])cc5)c5ccc(F)cc54)c(OCCOC(=O)CC4=C(C)/C(=C/c5ccc([S+](C)[O-])cc5)c5ccc(F)cc54)cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc2c1c(-c1ccc3occc(=O)c3c1)nn2C1CCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1CSC(NN=C2CCCCCC2)=N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CSCCNCc1ccc(-c2ccc3ncnc(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)c3c2)o1.Cc1ccc(S(=O)(=O)O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3cc(OCC4CCCO4)c4c(c23)OCCO4)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OCCCNCc1ccc(-c2cc3nccc(Nc4ccc5[nH]ccc5c4)c3s2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN[C@H]1C[C@@H]2O[C@](C)([C@H]1OC)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccccc3)c2cc1OC.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(C#CCN1CCOCC1)Nc1ccc2ncnc(Nc3ccc(F)c(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1nc(N(C)c2ccc(OC(C)C)cc2)c2cc(C)oc2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCc1cc(Nc2nccc(-c3c(-c4cccc(C(=O)Nc5c(F)cccc5F)c4)nc4ccccn34)n2)c(OC)cc1N1CCC(N2CCN(S(C)(=O)=O)CC2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)CCOc1ccc(/C=C/c2cc(-c3cc4c(=O)[nH]cnc4[nH]3)ccn2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(C(=O)N2CCOCC2)[nH]c1/C=C1\C(=O)Nc2ncnc(Nc3ccc4c(ccn4Cc4ccccc4)c3)c21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN(CC)[C@@](C)(C#Cc1ncnc2cc(OC)c(OC)cc12)CCc1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(CNc2nc(N)nc3[nH]c4ccc(O)cc4c23)cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2nccc(Nc3ccccc3-c3ccn(C)n3)n2)c(OC)cc1N1CCC(N(C)C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCNCc1cn2ncnc(Nc3ccc4c(cnn4Cc4ccccc4)c3)c2c1CC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1Cc2ccc(Oc3ccc(Nc4ncnc5ccn(CCO)c45)cc3Cl)cc2N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(-c2[nH]nc3ncnc(Nc4cccc(Cl)c4)c23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC1=C(C(=O)Nc2cccc([N+](=O)[O-])c2)C(c2ccc(O)cc2O)NC(NN2C(=O)/C(=C\c3cccs3)SC2=S)=N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/CNCC1COC1)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCOC(=O)Nc1ccc(Nc2ncnc3cc(OC)c(OC)cc23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncc([N+](=O)[O-])n1C/C(=N/NC(=O)c1ccc(N)cc1)c1ccc(Br)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)OC(Cc1c(SSc2[nH]c3ccccc3c2CC(OC(C)=O)C(=O)NCc2ccccc2)[nH]c2ccccc12)C(=O)NCc1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(Cl)cc3F)ncnc2cc1OCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN(CC)[C@@](C)(C#Cc1ncnc2cc(OC)c(OC)cc12)Cc1ccncc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1ccc(Cl)cc1)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc2c(Cc3cccc(Br)c3)ncnc2cc1OCCCn1ccnc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)n1nc(-c2ccc(C#N)c(O)c2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#C/C(=C\c1cc(O)c(O)c(Br)c1)S(=O)(=O)/C(C#N)=C/c1cc(O)c(O)c(Br)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc2cc(Nc3ccnc4cc(-c5ccc(CNCCOCCO)cc5)sc34)ccc2[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CCC(N2C(=O)N(Cc3ccccc3)Cc3cnc(Nc4ccc(N5CCN(C)CC5)cc4OC)nc32)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCC#CC(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(Nc2ncnc3ccc(NC(=O)NCCCl)cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc2cc(-c3nn(C(C)C)c4ncnc(N)c34)ccc2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CC[C@H](N2C(=O)N(c3cc(OC)ccc3F)Cc3cnc(Nc4ccc(N5CC6(CCN(C)CC6)C5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=FC(F)(F)c1cccc(Nc2ncnc3[nH]c4c(c23)CCCC4)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/c1cccs1)Nc1ccc2ncnc(Nc3cccc(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCC(c2c(O)cc(O)c3c(=O)cc(-c4ccccc4Cl)oc23)[C@@H](O)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(=O)CNc1cccc(Oc2cc(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)ncn2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)n1nc(-c2cccc(C#N)c2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(-c4nc5ccccc5s4)c(F)c3)c2cc1OCCCN1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(-c2cn(-c3ccccc3)c3ncnc(N)c23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC[N+](C)(C)[C@H]1C[C@@H]2O[C@](C)([C@H]1OC)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2n[nH]c3cc(-c4cncc5ccccc45)ccc23)c(OC)cc1N(C)CCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(-n2c3nc(Nc4ccc(N5CCN(NC)CC5)cc4OC)ncc3c(=O)n3ccnc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC1(C)OCc2c(Nc3n[nH]c(Cl)c3Cl)nc(-c3cn[nH]c3)nc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCN(c2ccc(NC(=O)c3c(Nc4ccccc4Cl)cc[nH]c3=O)cc2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=COC(=O)N(C)/N=N/c1ccc2ncnc(Nc3cccc(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)CCOc1ccc(Nc2ncc3nc(Sc4ccc(F)cc4)n(C4CCOCC4)c3n2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CC[C@H](N2C(=O)N(c3ccc(F)cc3)Cc3cnc(Nc4ccc(N5CCN(C)CC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc2c1c(-c1ccc(F)c(O)c1)nn2[C@@H]1CCNC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=c1c(-c2cccc(Cl)c2)coc2cc(O)cc(O)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CCC[C@@H](n2nc(C3CC3)c3c(N)ncnc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC(=O)Nc1ccc(Nc2ncnc3cc(OC)c(OC)cc23)cc1C.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C(=O)NS(=O)(=O)c2ccc(Br)cc2)c(Cl)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncnc(Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)c1/C=C/c1ccc(CNCCS(C)(=O)=O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CC[C@@H](N2C(=O)N(Cc3ccccc3)Cc3cnc(Nc4ccc(N5CCN(C)CC5)cc4OC)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNC(=O)COc1cccc(Nc2ncc(F)c(Nc3ccc4c(c3)NC(=O)C(F)(F)O4)n2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN1CCN(c2ccc(Nc3nccc(-c4c(-c5cccc(NC(=O)Cc6ccccc6)c5)nc5sccn45)n3)cc2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cn1nc(-c2ccc(Br)c(O)c2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1[nH]c(/C=C2\C(=O)Nc3ccc(C(=O)NNc4ccccc4)cc32)c(C)c1CCC(=O)O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1NC(=O)/C=C/CNC1CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(Oc2nc(Nc3n[nH]c4ccccc34)cc(N3CCN(C(C)=O)CC3)n2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(Oc2ccc(Nc3ncnc4[nH]nc(OCCN5CCC(O)CC5)c34)cc2F)cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CCn1nc(-c2ccc(OCC)c(OC)c2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COCCOc1cc2ncnc(Nc3cc(OC)c(OC)c(OC)c3)c2cc1OCCOC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1ccc(Nc2ncnc3ccc(-c4cnn(C5CCNCC5)c4)cc23)cc1Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CO[C@H]1[C@@H](N(C)C(=O)C(F)(F)F)C[C@@H]2O[C@@]1(C)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCN1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OCCn1ccc2ncnc(Nc3ccc(Oc4cccc5[nH]ccc45)c(Cl)c3)c21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Clc1ccc(Nc2nnc(Cc3ccncc3)c3ccccc23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cccc(-c2cc3c(N[C@H](C)c4ccccc4)ncnc3s2)c1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1nc(Nc2ccc(Cl)cc2F)c2cc(CCc3ccccc3)[nH]c2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cccc(-c2cn(C3CC(C(=O)NCCO)C3)c3ncnc(N)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)C/C=C/C(=O)Nc1ccc2ncnc(Nc3cccc(I)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)O[C@H]1CN[C@H](C#Cc2cc3ncnc(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)c3s2)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1nccs1)[C@@H](c1cc(F)ccc1O)N1Cc2ccccc2C1=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNc1cc2c(Nc3cccc(Br)c3)ncnc2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1CSC(N/N=C/c2cccc(O)c2)=N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)C1CCN(c2ccc(Nc3cc4c(N5CCC(CO)CC5)nc(Nc5ccc(F)cc5)nc4cn3)nc2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(N2CCN(C)CC2)ccc1Nc1ncc(Cl)c(Oc2ccc(NC(=O)CO)cc2)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCN(/N=C/c2c(N)ncnc2Nc2ccc3c(cnn3Cc3cccc(F)c3)c2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCN(CCO)CCO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccccc1/C=C/C(=N/NC(N)=S)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1nccs1)Nc1cc(-c2cccnc2)ccc1OC(F)(F)F>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CN1CCC(CCO)CC1)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cn1ncc([N+](=O)[O-])c1C[N+](C)(C)C/C=C/C(=O)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1.[Br-]>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NCCCSc1nc(-c2ccc(F)cc2)c(-c2ccnc(Nc3ccccc3)c2)[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)Nc1cc2c(Nc3ccc(NS(=O)(=O)c4ccccc4)nc3)ncnc2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CCC1C(S)=Nc2ccccc21)NCc1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#CC1=C(N)N(c2cccnc2)C2=C(C(=O)CCC2)C1c1cc2ccccc2n2nnnc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCCCCNC(=O)COc1cc(O)c2c(=O)cc(-c3ccccc3)oc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1[nH]c2ncnc(NCc3ccccc3)c2c1C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1CSC(N2N=C(c3ccc(Cl)cc3)CC2c2ccc(Cl)cc2)=N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C2=NN(C3=NC(c4ccccc4)CS3)C(c3cccc4ccccc34)C2)cc1C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Oc1ccc(CN(Cc2cc(Br)cc(Br)c2O)C(=S)Nc2ccccc2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COCCOc1cc2ncnc(Nc3cccc(NC(=O)C45CC6CC(CC(C6)C4)C5)c3)c2cc1OCCOC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OC[C@H](O)CN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cn1c(=O)c(-c2c(Cl)cccc2Cl)cc2cnc(Nc3cccc(CO)c3)nc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC1(C)OCc2c(Nc3nsc4ccccc34)nc(-c3cn[nH]c3)nc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccccc1CNc1nc(N)nc2[nH]c3cc(C)c(O)cc3c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc2c(c1)Oc1ncnc(Nc3ccc(F)c(Cl)c3)c1NC2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(Br)c(Br)c(/C=N/c2ccc3[nH]c(=O)[nH]c3c2)c1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(Nc2ncc3c(n2)-c2ccc(Cl)cc2SC3)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc2ncnc(Nc3cccc(-c4nc[nH]n4)c3)c2cc1OCC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COCCNC(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1O[C@H]1CCOC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(/C=C(\C#N)C(N)=O)cc(SCCCC(=O)O)c1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC1CCN(c2nccc(Nc3cc4c(ccc(=O)n4C(C)C)cn3)n2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(CN(Cc2ccc(F)cc2)C(=O)Nc2ccccc2)c1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCCCOc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/c1cccc(-c2ccc3ncnc(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)c3c2)c1)NO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cccc(-c2cn(C3CCN(CC(=O)N(C)C)CC3)c3ncnc(N)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#Cc1cnc2cc(OC[C@H](O)CO)c(NC(=O)C3CSSC3)cc2c1Nc1ccc(OCc2cccc(F)c2)c(Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(-n2c(=O)cnc3cnc(Nc4ccc(N5CCN(C)CC5)cc4)nc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc2ncnc(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)c2cc1NC(=O)[C@H]1COC(=O)N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(C(=O)NCCN(C)C)ccc1-c1cc2c(N[C@H](C)c3ccccc3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(N(CCCl)CCN/N=N/c2ccc3ncnc(Nc4cccc(Cl)c4)c3c2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)ncn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1nc2c(Cc3ccc(F)c(Cl)c3)ncnc2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCS(=O)(=O)CC(=O)NCCn1ccc2ncnc(Nc3ccc(Oc4cccc(C(F)(F)F)c4)c(Cl)c3)c21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(F)ccc1-c1cc2c(N[C@H](C)c3ccccc3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNC(=O)[C@@H](C)N(C)Cc1cc2c(Nc3cccc(Cl)c3F)ncnc2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC(=O)CN(C)[C@H]1C[C@@H]2O[C@](C)([C@H]1OC)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC1(C)OCc2c(Nc3n[nH]c4c(Cl)cc(F)cc34)nc(-c3cn[nH]c3)nc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COCCN(CCOC)S(=O)(=O)c1ccc(Nc2nc3ncnc(Nc4ccc(F)c(Cl)c4)c3s2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)C/C=C/C(=O)NCCSc1nc(-c2ccc(F)cc2)c(-c2ccnc(Nc3ccccc3)c2)[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1ccncc1)Nc1cc(-c2cccnc2)ccc1OC(F)(F)F>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc2c1c(-c1ccc3cn[nH]c3c1)nn2C1CCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1ccc2ncnc(Nc3ccc(S(=O)(=O)Nc4nccs4)cc3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)c1ccc(Nc2[nH]nc3ncnc(Nc4cccc(Cl)c4)c23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(N/N=C/c1cccs1)N1CCc2ncnc(Nc3ccc(F)c(Cl)c3)c2C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(Nc2cc(NC(=O)c3ccccc3)ncn2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1ccc(-c2nc(-c3ccccc3)[nH]c2-c2ccnc3[nH]ccc23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC[C@@H](Nc1ncnc2oc(-c3ccc(OC)cc3)cc12)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1N=C(N2CCC[C@H]2C(=O)Nc2ccc3ncnc(Nc4cccc(Cl)c4)c3c2)S/C1=C/c1cccnc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CSc1cn2c(-c3cn[nH]c3)cnc2c(Nc2cc(C)ns2)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(C)cc(-n2c(=O)cc(C)c3cnc(Nc4ccc(N5CCN(C)CC5)cc4OC)nc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CC[C@H](Nc2nc(Nc3ccccc3)nc3cnc(Nc4ccc(N5CCN(C)CC5)cn4)cc23)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3ccc(OCCCCNCCS(C)(=O)=O)cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCN(c2ccc(Nc3ncc4nc(Sc5ccccc5)n([C@H]5CC[C@H](O)CC5)c4n3)cc2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1ccc(Nc2cc(NC(=O)c3ccccc3)ncn2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#CC(C#N)=C(C#N)c1cc(O)c(O)c(O)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1ccc(F)cc1)c1ccc(N(CCCl)CCCl)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(N2CCN(C)CC2)ccc1Nc1ncc(Cl)c(Oc2cccc(NC(=O)CCN3CCCCC3)c2)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(C)c1Nc1ncc(-c2ccc(OCC3CCCN(C)C3)cc2)n2cncc12.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1CSC(N2N=C(c3ccc(Br)cc3)CC2c2ccc(Br)cc2)=N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NCCOCc1ccn2ncnc(Nc3ccc4c(cnn4Cc4cccc(F)c4)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(-c2nn(C3CNC3)c3ncnc(N)c23)cc1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)CCCNC/C=C/C(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCc1c(C(=O)NC)cn2ncnc(Nc3ccc4c(cnn4Cc4ccccc4)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCCCCn1ccnc1[N+](=O)[O-]>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1C(=O)C(=C(C#N)C#N)c2cc(O)ccc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(CC(O)C(O)C(O)C(O)CO)c1cc2ncnc(Nc3cccc(Br)c3)c2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cccc(Nc2ncnn3ccc(CN4CC[C@@H](N)[C@H](O)C4)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=c1csc(-c2nnc(Cc3nc4ccccc4[nH]3)o2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1ccc(-c2ncn(CCN3CCOCC3)c2-c2ccc3[nH]ncc3c2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COCOc1cc(-c2nn(C(C)C)c3ncnc(N)c23)ccc1Br>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(O[C@H]1CN[C@H](C#Cc2cc3ncnc(Nc4ccc5c(ccn5CCc5ccccc5)c4)c3s2)C1)N1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNC(=O)c1ccc(Sc2cccc(NC(=S)Nc3ccc(Cl)c(C(F)(F)F)c3)c2)nc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc2c(c1)CNc1c(NCc3ccccc3)ncnc1O2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(C=C(C#N)C#N)cc(C)c1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCc1c(C(=O)NCCCn2ccnc2)cn2ncnc(Nc3ccc4c(cnn4Cc4ccccc4)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(C)c1-c1cc(C)c2nc(Nc3ccc(OCCN4CCCC4)cc3)nnc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC1(C)OCc2c(Nc3n[nH]c4c(C(F)(F)F)cccc34)nc(-c3cn[nH]c3)nc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cn1c(=O)c2[nH]c(SC/C(=N/O)c3ccc(Br)cc3)nc2n(C)c1=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(-c2csc(=N)n2CC(=O)N2CCOCC2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cl.O=[N+]([O-])c1ccc2sc3c(Nc4cccc(Br)c4)ncnc3c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc2c(Nc3cccc(Br)c3)ncnc2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc2ncn(CC(=O)c3c[nH]c4ccccc34)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(NCCc1ccc(O)c(Br)c1)/C(Cc1cc(Br)c(O)c(-c2cc(C/C(=N/O)C(=O)NCCc3ccc(O)c(Br)c3)cc(Br)c2O)c1)=N\O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OCCN(CCO)CCCNc1cc2ncnc(Nc3cccc(Br)c3)c2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1nc2cnc(Nc3ccnc(N4CCC(C)(C(N)=O)C4)n3)cc2n1C(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)C1CCN(c2ccc(Nc3cc4c(N5CCOCC5)nc(Nc5ccc(F)cc5)nc4cn3)nc2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCOC(=O)Nc1ccc(Nc2ncnc3cc(OC)c(OC)cc23)cc1C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(Nc2ncnn3ccc(CN4CCC(N)CC4)c23)cc1Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Clc1cc(Nc2ncnc3[nH]nc(OCCN4CCCC4)c23)ccc1OCc1ccccn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@@H](Nc1ncnc2sc(Br)cc12)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CC[C@H](N2C(=O)N(c3cccc(OC(F)(F)F)c3)Cc3cnc(Nc4ccc(N5CCN(C)CC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C1=CC2=C(Cc3nnc(Cc4nc5ccccc5[nH]4)o3)CNC2C=C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1nc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1OCCCN1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)CCCNC(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1O[C@H]1CCOC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncnc(Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)c1/C=C/c1ccc(CNC(C)C)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1ccc2ncnc(Nc3ccc(S(N)(=O)=O)cc3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1ccc(OC)c(Nc2cc(-c3nc(SC)n(C)c3-c3ccc(F)cc3)ccn2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)NCc1cccc(-c2nc(-c3ccc(F)cc3)c(-c3ccnc4[nH]c(-c5ccccc5)cc34)[nH]2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C/N=N/Nc1ccc2ncnc(NCc3ccccc3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Oc1cccc(Nc2ccnc3[nH]c4ccccc4c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1OCC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@@H](COc1cccc2ncnc(Nc3ccc(OCc4ccccn4)c(Cl)c3)c12)N(C)C(=O)CO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(Nc2ncnc3[nH]c(C)c(C)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3cc4oc(=O)n(CCCN5CCOCC5)c4cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNC(=O)c1nc(-c2ccc(Cl)c(S(=O)(=O)Nc3cccc(F)c3C)c2)cnc1N>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCCCCCNC(=O)COc1cc(O)c2c(=O)cc(-c3ccccc3)oc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(S(=O)(=O)NC(=O)c2ccc(C)nc2Cl)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCNc1ncc(C(=O)Nc2cc(C(=O)Nc3ccc(CN4CCN(C)CC4)c(C(F)(F)F)c3)ccc2C)cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@H](Nc1ncnc2c1sc1ccccc12)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(cc1OC)Nc1ncn(C)c(=O)c1C2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(Nc2nc(Nc3ccc(CN4CCOCC4)cc3OC)ncc2Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CCc1ccc(O)cc1)c1c(O)cc(O)cc1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCc1ccc(Nc2ncnc3[nH]c(C)c(C)c23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2n[nH]c3cc(-c4ccc(F)cc4)ccc23)c(OC)cc1N(C)CCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1ccc2cc(-c3nn(C(C)C)c4ncnc(N)c34)ccc2c1.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC/C=C\C(=O)Nc1cc(Nc2nc(C)cc(-c3cn(C)c4ccc(F)cc34)n2)c(OC)cc1N(C)CCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(Nc2ncnc3ccncc23)c1NCCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC1CCCCN1C/C=C/C(=O)N1CCOc2cc3ncnc(Nc4ccc(F)c(Cl)c4)c3cc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCNc1ncc2cc(-c3c(Cl)cccc3Cl)c(=O)n(C)c2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)n1nc(-c2ccc(O)cc2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(-c2nn(C[C@H](C)CO)c3ncnc(N)c23)cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#Cc1cc2c(N)ncnc2nc1NCCO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(O)c1ccccc1-c1nnc(Cc2nc3ccccc3[nH]2)o1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cccc(-c2cn(C3CCC3)c3ncnc(N)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(-c3c[nH]c4ccc(Br)cc34)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(cc1O)NC(=O)C2=C(C#N)C#N>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1nc(Nc2cccc(Br)c2)c2cc(Cc3ccc(-c4ccccc4)cc3)[nH]c2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)c1cccc(-c2nn(C(C)C)c3ncnc(N)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1ccc(Nc2ncnc3ccc(C#Cc4cccs4)cc23)cc1Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCN(CC#CC(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cn1)CCCC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(/C=C(\C#N)C(=O)O)ccc1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Brc1cccc(Nc2[nH]cnc3nc4ccccc4c2-3)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(NC(=O)Nc4ccc(C)c(C)c4)c(Cl)c3)ncnc2cc1OCCN1CCCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncc(C#N)c(Nc3cc(Cl)c(O)c(Cl)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(Nc2ncnc3[nH]nc(OCCN4CCC(O)CC4)c23)ccc1OCc1cccc(F)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=FC(F)(F)c1cccc(Nc2ncnc3cc(OCCCN4CCOCC4)c4c(c23)OCCO4)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(NC3CC3c3ccccc3)c2cc1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccccc1-c1cc2c(NCc3cccs3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1c(Br)cccc1Nc1ncnc2ccncc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(O[C@H]1CN[C@H](C#Cc2cc3ncnc(Nc4ccc(OCc5ccccn5)c(Cl)c4)c3s2)C1)N1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C2=NN(C(N)=S)C(c3ccc(O)cc3)C2)cc1C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CCN(c2ccc(Nc3ncc(Cl)c(Oc4cccc(COc5no[n+]([O-])c5S(=O)(=O)c5ccccc5)c4)n3)c(OC)c2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CCC[C@@H](n2c(=O)c(-c3ccccc3Cl)c(C)c3cnc(Nc4ccc(N5CCN(C)CC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=c1oc2cc3ncnc(Nc4ccc(OCc5ccccn5)cc4)c3cc2n1CCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CCC[C@H]1C(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3F)ncnc2cc1OCCOC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#CC(C#N)=Cc1ccc(C=O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O[C@H]1CC[C@H](Nc2ncc3nc(Nc4c(F)cc(F)cc4F)n([C@H]4CCOC4)c3n2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)C/C=C/C(=O)N1CCc2c(sc3ncnc(NCCO)c23)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC/C=C\C(=O)Nc1cc(Nc2nc(C)cc(-c3cn(C)c4ccc(F)cc34)n2)c(OC)cc1N(C)CCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(C)Oc1cc2ncnc(Nc3ccc(F)c(C#N)c3)c2c2c1OCCO2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1ccc(Nc2ncnc3sc4c(c23)CCCC4)cc1)Nc1cccc(Br)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COCC(=O)Nc1ccc2ncnc(Nc3cccc(I)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Oc3ccc(C4SCC(=O)N4NC(=O)Nc4ccc(F)c(F)c4F)cc3)ccnc2cc1OCCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)n1nc(-c2cc(O)cc(F)c2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1NCc2cccc(Oc3ccc(Nc4ncnc5ccn(CCO)c45)cc3Cl)c21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1B1OC(C)(C)C(C)(C)O1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCCCCCCNC(=O)COc1cc(O)c2c(=O)cc(-c3ccccc3)oc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cn1c(SSc2c(C(=O)Nc3ccccc3)c3cccnc3n2C)c(C(=O)Nc2ccccc2)c2cccnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#CC(C#N)=Cc1ccc(NC(=O)CCC(=O)O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)n1nc(-c2cc3ccccc3nc2Cl)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(N2C(=O)C(Cc3ccc(F)cc3)N(C)C(=O)c3cnc(Nc4ccc(N5CCN(C)CC5)cc4OC)nc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/CNCc1ncccn1)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1nc2c(c(Nc3ccc(F)cc3)n1)-c1ccccc1C2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1ccc(Nc2ncnc3nc(Nc4ccc(CN5CCCCC5)cc4)sc23)cc1Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(-n2c(=O)cc(C)c3cnc(Nc4ccc(N5CCN(C)CC5)cc4OCCC)nc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C2=NN(C3=NC(=O)CS3)C(c3ccc(Cl)cc3)C2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2nc(Cl)nc(Nc3ccc(S(=O)(=O)Nc4nccs4)cc3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(-c2nc3ccccc3s2)ccc1Nc1ncnc2cc(OC)c(OC)cc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@@H](c1ccccc1)n1ncc2cc(Nc3ncnn4ccc(COC[C@@H]5CNCCO5)c34)ccc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=c1ccc(CCNc2ccnc3oc4ccccc4c23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1nc2cnc(Nc3ccnc(N4CC[C@H](O)C(F)(F)C4)n3)cc2n1C(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CS(=O)(=O)N1CCC[C@@H]1Cn1cc(-c2ccc3c(c2)OCO3)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc2c1c(-c1ccc3[nH]c(=O)ccc3c1)nn2C1CCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=C=CCCCOc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(OC2CCOC2)c2c(Nc3ccc(F)c(Cl)c3)ncnc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)CCCCNc1c(Br)cccc1Nc1ncnc2ccncc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(N2CCN(C)CC2)ccc1Nc1ncc(Cl)c(Oc2ccc(N=[N+]=[N-])cc2)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Oc3ccc(NS(=O)(=O)c4cc(-c5ccsc5)ccc4F)cc3F)ccnc2cc1OCCCN1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC(=O)c1ccc(CNC(=O)c2c(SSc3c(C(=O)NCc4ccc(C(=O)OC)cc4)c4ccccc4n3C)n(C)c3ccccc23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cn1c(SSc2c(C(=O)NCC(=O)O)c3ccccc3n2C)c(C(=O)NCC(=O)O)c2ccccc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1C(=S)C(C(=O)Nc2ccccc2)c2ccccc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C(=O)Nc2ccc(CN3CCN(C)CC3)c(C(F)(F)F)c2)cc1NC(=O)c1cnc2[nH]ncc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncnc(Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)c1C#Cc1ccc(CN2CCOCC2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC(=O)CNC(=O)c1ccc(Nc2ncnc3cc(OCCN4CCCCC4)c(OC)cc23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(Nc2ncnc3ccc(NC(=O)N(C)CCCl)cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OCC(O)CNc1c(Br)cccc1Nc1ncnc2ccncc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cccc(-c2cn(C3CCN(CCN(C)CCO)CC3)c3ncnc(N)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNCC(O)COc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1ccc(C2=NN(c3ccccc3)C(c3cccc4ccccc34)C2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(O)CCc1c(SSc2[nH]c3ccccc3c2CCC(=O)O)[nH]c2ccccc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(/C=C(\C#N)C(N)=O)cc(CSc2ccc(O)cc2)c1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc2c(Nc3cccc(Br)c3)ncnc2cc1OCCCN1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#C/C(=C/c1cc(O)ccc1O)C(=O)O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc2ncc(C#N)c(N[C@@H]3C[C@H]3c3ccccc3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(=O)C1=C(N)N(c2cccnc2)C2=C(C(=O)CCC2)C1c1cc2cc(C)ccc2n2nnnc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(NC(=O)Nc2nccs2)cc(-c2cn(C)cn2)c1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(C)Oc1cc2ncnc(Nc3ccc(C)c([N+](=O)[O-])c3)c2c2c1OCCO2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(NCCCN1CCOC1=O)c1cnc(NCc2cc(Cl)ccc2Cl)nc1NC1CCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3cccc(F)c3)ncnc2cc1OCCOCCNC(=O)Cn1cnc([N+](=O)[O-])n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccccc1-c1cc2c(NCc3ccncc3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1c(C(=O)NCCCN2CCN(C)CC2)[nH]c2cnnc(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NS(=O)(=O)c1ccc(Nc2nc3ncnc(Nc4ccc(F)c(Cl)c4)c3s2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OCCn1ccc2ncnc(Nc3ccc(Oc4cccc5[nH]ncc45)c(Cl)c3)c21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCc1c(NC(=O)OCCn2ccnc2)cn2ncnc(Nc3ccc4c(cnn4Cc4ccccc4)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CCl)OCCn1c(=O)oc2cc3ncnc(Nc4ccc(F)c(Cl)c4)c3cc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Brc1cccc(Nc2ncnc3cc4c(cc23)OCCCSCCCO4)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(-c2n[nH]c3ncnc(N)c23)cc1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1O[C@H]1CCOC1)N1CCCC(F)(F)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(OC)cc(-c2cc3cnc(N)nc3nc2NC(=O)NC(C)(C)C)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#CC(C#N)=C(C#N)c1ccc(O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)Nc1cc2c(Nc3ccc4c(c3)CCN4Cc3ccccn3)ncnc2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CCC[C@@H]1COc1nc(Nc2ccc(N3CCN(C(C)=O)CC3)cc2)nc2[nH]cc(-c3cncc(NC)n3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCCCC(=O)ON[C@H](CS(C)(=O)=O)c1ccc(-c2ccc3ncnc(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)c3c2)o1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(N2CCN(C)CC2)ccc1Nc1ncc(Cl)c(Oc2ccccc2N=C=S)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=O)COCc1ccn2ncnc(Nc3ccc4c(cnn4Cc4cccc(F)c4)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1cccc(-c2cc(=O)c3c(N)c(O)c(N)cc3o2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)Cc1ccc(-c2cc3c(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)ncnc3s2)[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(NCCCN1CCCC1=O)c1cnc(NCc2cc(Cl)ccc2Cl)nc1NC1CCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)=CC[C@@H](OC(=O)/C=C\C(=O)c1ccc(C)cc1)C1=CC(=O)c2c(O)ccc(O)c2C1=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(C#N)cnc(Nc3cccc(Br)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Oc1nnc(-c2c[nH]c(-c3ccccc3)c2-c2ccccc2)c2cn(-c3ccc(Cl)cc3)nc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(-n2c(=O)cnc3cnc(Nc4ccc(N5CCN(C)CC5)cc4OC)nc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)N1CCN(Cc2ccc(-c3ccc4ncnc(Nc5ccc(OCc6cccc(F)c6)c(Cl)c5)c4c3)o2)CC1.Cc1ccc(S(=O)(=O)O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncc(C#N)c(Nc3cccc(SC)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(NC(=O)c4ccccc4Cl)cc3)ncnc2cc1OCc1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCNC(=O)O[C@@H]1CN[C@@H](C#Cc2cc3ncnc(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)c3s2)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC/C=C/C(=O)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1ccc(-c2ncn(C3CCN(Cc4ccccc4)C3)c2-c2ccc3[nH]ncc3c2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(Nc2cc3c(cc2Nc2ccccc2)C(=O)NC3=O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(-c2nn(C[C@@H](C)CO)c3ncnc(N)c23)cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C2CC(c3ccc(Cl)c(Cl)c3)=NN2C(N)=S)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CC[C@@H](Nc2nc(Nc3ccc(N4CCN(C)CC4)cc3)c3ncn(C(C)C)c3n2)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(c1cccc(Br)c1)c1nc(N)nc2[nH]c(Cc3ccccc3)cc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(-c2nn(C3CCCC3)c3ncnc(N)c23)cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(-c2c3c4cc(OCCN(C)C)c(OCCN(C)C)cc4oc(=O)c3n3ccc4cc(O)c(OC)cc4c23)cc1O.O=C(O)C(F)(F)F.O=C(O)C(F)(F)F>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#CCCn1cc(Nc2nc(OC[C@H]3CN(C(=O)C=C)C[C@@H]3OC)c3c(Cl)c[nH]c3n2)cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(C2=NN(C3=NC(=O)CS3)C(c3ccc4ccccc4c3)C2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(/N=C/c2c(N)ncnc2Nc2ccc3c(cnn3Cc3cccc(F)c3)c2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1c(C=O)cccc1-c1cc2c(N[C@H](C)c3ccccc3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)N1CCN(C(=O)Cn2c(-c3ccc(Cl)cc3)csc2=N)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(/C=C/CCc3ccccc3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=S)N1N=C(c2ccc(Cl)c(Cl)c2)CC1c1ccc(F)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(N3CCc4ccccc43)c2cc1NC(=O)/C=C/CN1CCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#Cc1ccc(-c2csc(N3N=C(c4ccc(N5CCCCC5)cc4)CC3c3ccc(Cl)cc3)n2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Oc3cccc(Br)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2nccc(Nc3ccccc3-c3ccn(C)n3)n2)c(OC)cc1N(C)CCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(C(=O)Nc2nc3cccc(C)c3n2[C@@H]2CCCCN(C(=O)/C=C/CN(C)C)C2)ccn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#C/C(=C\c1ccc(O)c(O)c1)C(=O)NCc1cccc(CNC(=O)/C(C#N)=C/c2ccc(O)c(O)c2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(CO)cc1-c1cc2c(N[C@H](C)c3ccccc3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)NC1=CC=C2N=CN=C(Nc3cccc(Cl)c3)C21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)=CC[C@@H](OC(=O)/C=C\C(=O)c1ccc(C)cc1Cl)C1=CC(=O)c2c(O)ccc(O)c2C1=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2nccc(Nc3ccccc3-n3cccn3)n2)c(OC)cc1N(C)CCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(/C(CSc2nc3c([nH]2)c(=O)n(C)c(=O)n3C)=N\O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2nccc(Nc3ccc(F)c(Cl)c3)c2cc1NC(=O)/C=C/C(C)N(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncc(C#N)c(Nc3cccc(Cl)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCNC(=O)Nc1ccc2ncnc(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc(N)c(C(=O)Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)cc1NC(=O)c1ccc(CN2CCN(C)CC2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(C)c1-c1cc2cnc(N)nc2nc1NC(=O)NC(C)(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(O)CNc1cc2ncnc(Nc3cccc(Br)c3)c2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncc(C#N)c(N[C@@H]3C[C@H]3c3ccccc3)c2cc1OCCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(C(=O)N(C)[C@H]2C[C@@H]3O[C@](C)([C@H]2OC)n2c4ccccc4c4c5c(c6c7ccccc7n3c6c42)C(=O)NC5)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cccc(-c2ccc3c(c2)NC(=O)/C3=C\c2[nH]c3c(c2CCC(=O)O)CCCC3)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cc(Nc2ncnc3cc(OC)c(OC)cc23)ccc1F>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN/N=C/c1c(N)ncnc1Nc1ccc2c(cnn2Cc2cccc(F)c2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/C(=O)NCCCn1ccnc1)Cc1cc2c(Nc3cccc(Br)c3)ncnc2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCC(O)CN(C)[C@H]1C[C@@H]2O[C@](C)([C@H]1OC)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(=O)n2nc(SCc3ccc(Cl)cc3Cl)nc2[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3cccc(Cl)c3F)c2cc1CN1CC(C(N)=O)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCOC(=O)Nc1ccc(Nc2ncnc3cc(OC)c(OC)cc23)cc1Cl.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(-c4nc5ccccc5s4)cc3O)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3cc(O[C@H]4CCOC4)c(NC(=O)/C=C/CN4CCCCC4)cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCN(C(=O)Cn2c(-c3ccc(Cl)cc3)csc2=N)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cn1ncc2cc(Nc3ncnn4ccc(CN5CCC(N)CC5)c34)ccc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(-c2nnc(Cc3nc4ccccc4[nH]3)o2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1cccc2ncnc(Nc3cccc(Br)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1nc(Nc2cccc(F)c2)c2cc(Cc3ccc(Cl)cc3Cl)[nH]c2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1ccc(Nc2ncnc3nn4ccccc4c23)cc1Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1ccc2ncnc(Nc3cccc(Cl)c3)c2c1)C1CCC2(CC1)OCC1(OO2)C2CC3CC(C2)CC1C3>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=S(=O)(Nc1ncccn1)c1ccc(Nc2nncc3ccccc23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1nc2cnc(Nc3ccnc(OCC(C)(C)C(N)=O)n3)cc2n1C(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc2c(c1)CNc1c(Nc3ccc(Cl)cc3)ncnc1O2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(=O)Nc1cc(Nc2ncnc3cc(OC)c(OC)cc23)ccc1C.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1ccccc1NC(=O)/C=C/c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccccc1C(=Cc1coc2nc(=N)[nH]c(N)c12)C(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc2c(c1)CCCN2c1nc(C)nc2oc(C)cc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(Oc2ccc(Nc3ncnc4[nH]nc(OCCN5CCC(O)CC5)c34)cc2C)cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(N2CCN(C)CC2)ccc1Nc1ncc(Cl)c(Oc2ccccc2N=[N+]=[N-])n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCNCc1ccc(-c2cc3ncnc(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)c3s2)[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/CC(=O)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1)Nc1ccc2c(Nc3cccc(Br)c3)ncnc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCCOP(N)(=O)N(CCCl)CCCl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNc1cc2ncnc(Nc3cccc(Br)c3)c2cc1N>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C2=NN(C(N)=S)C(c3ccccc3F)C2)cc1C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)Nc1cc2c(Nc3ccc(Cc4ccccn4)cc3)ncnc2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OCc1cccc(-c2cc3c(N[C@H](CO)c4ccccc4)ncnc3s2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(Cl)cc3F)ncnc2cc1OCC1CCNCC1.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CCc1c(SSc2[nH]c3ccccc3c2CCC(=O)NCCc2ccccc2)[nH]c2ccccc12)NCCc1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(OCc4cccc(F)c4)c(Cl)c3)ncnc2cc1OCCCCn1ccnc1[N+](=O)[O-]>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCN(C/C=C/C(=O)Nc2cc3c(Nc4ccc(Cc5ccccn5)cc4)ncnc3cn2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C2CC(c3ccc(C)c(C)c3)=NN2C(N)=S)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(Nc2c(C#N)cnc3cc(OC)c(OC)cc23)cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC1CCN(c2nccc(Nc3cc4c(cn3)ncn4C3CCCC3)n2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CN1CCOCC1)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CO/N=C/c1c(N)ncnc1NCc1ccc(F)c(Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)CCCCNc1cc2ncnc(Nc3cccc(Br)c3)c2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(-n2c(=O)cnc3cnc(Nc4ccc(NC(C)=O)cc4)nc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(Nc2ncnc3cc4c(cc23)N(CCCN2CCOCC2)C(=O)CO4)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(/C=C2\CCC/C(=C\C3CCCCC3)C2=O)cc(OC)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1C[C@H](F)[C@@H](COc2nc(Nc3cnn(C)c3)nc3[nH]cc(Cl)c23)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1ccc(OC)c(Nc2cc(-c3[nH]c(SC)nc3-c3ccc(F)cc3)ccn2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(-c2cc3c(NCc4ccccc4)ncnc3o2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/CN1CCOCC1)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccccc3Br)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1)c1cccc2c1OCCO2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1ccc(-c2nn(C(C)C)c3ncnc(N)c23)cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)cc3)c2cc1OC.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(Nc2ncnc3cnc(N(C)C)cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(/C=C(\C#N)C(N)=O)cc(Sc2nc3ccccc3s2)c1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCN(CCOc2n[nH]c3ncnc(Nc4ccc(OCc5ccccn5)c(Cl)c4)c23)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCNc1ncc2cc(-c3c(Cl)cccc3Cl)c(=O)n(C)c2n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CO[C@H]1[C@@H](N(C)C(=O)CCC(=O)O)C[C@@H]2O[C@@]1(C)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ccc2ncnc(Nc3cccc(C(F)(F)F)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(-n2nc(C(C)(C)C)cc2NC(=O)Nc2ccc(OCCN3CCOCC3)c3ccccc23)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccccc1Cc1cc2c(N(C)c3cccc(Br)c3)nc(N)nc2n1C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Oc1ccc2ncnc(Nc3ccc(OCc4ccccc4)cc3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N(CCCN1CCOCC1)c1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(Nc2ncnc3cnc(N(C)C)nc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(C)c(C)c(-c2cc3cnc(NCCCN4CCN(C)CC4)nc3nc2NC(=O)NC(C)(C)C)c1C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COCOCC#CC(=O)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=O)CCC1C(S)=Nc2ccccc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COCCOc1cc2ncnc(Nc3ccc(F)c(C#N)c3)c2c2c1OCCO2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)CCNCc1ccc(-c2ccc3ncnc(Nc4ccc(OCc5ccccc5)c(C(F)(F)F)c4)c3c2)o1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(-n2c(SCC(=O)Nc3ccccc3Cl)nc3sc(C)c(C)c3c2=O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1CSC(N2N=C(c3ccc(F)cc3)CC2c2ccc(F)cc2)=N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(C)cc3F)ncnc2cc1OCC1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CNC(=S)N(C)[C@H]1C[C@@H]2O[C@](C)([C@H]1OC)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1CSC(N2N=C(c3ccncc3)CC2c2ccc(Br)cc2)=N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCN1CCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc2c1c(-c1ccc(F)c(O)c1)nn2[C@H]1CCOC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(/N=N/c3ccc(F)c(Cl)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N[C@@H]1CN(c2nc(Nc3cn(C)nc3OC)c3ncn(C(C)(C)C)c3n2)C[C@H]1F>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=C(CN1CCOCC1)C(=O)c1ccc(OCc2ccccc2)cc1.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3cc(O)c(Cl)cc3F)ncnc2cc1OCC1CCN(C)CC1.Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Cc1ccc(Cl)cc1)NS(=O)(=O)c1ccc(Br)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccccc1Nc1cc[nH]c(=O)c1C(=O)Nc1ccc(N2CCC(C)CC2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)CCNCc1nc(-c2ccc3ncnc(Nc4ccc5c(cnn5Cc5ccccc5)c4)c3c2)cs1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cn1c(=O)c(-c2c(Cl)cccc2Cl)cc2cnc(Nc3ccccc3)nc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2ncc(Cl)c(-c3cnn4ccccc34)n2)c(OC)cc1N1CC[C@@H](N(C)C)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1OCCCCCCn1ccnc1[N+](=O)[O-]>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CC[C@@H](n2c(=O)c(-c3ccccc3Cl)c(C)c3cnc(Nc4ccc(N5CCN(C)CC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CO[C@@H]1[C@H](N(C)C(=O)c2ccccc2)C[C@H]2O[C@]1(C)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)N[C@H]4O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1cccc(COc2ccc(Nc3cc(-c4ccccc4)ncn3)cc2Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(O)C1CCN(Cc2ccn3ncnc(Nc4ccc5c(cnn5Cc5cccc(F)c5)c4)c23)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(NS(=O)(=O)c1ccc(Br)cc1)c1cccnc1Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncc([N+](=O)[O-])n1C/C(=N/NC(=O)c1ccc(Cl)cc1)c1ccc(Br)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN(CC)C/C=C/C(=O)N1CCCOc2cc3ncnc(Nc4ccc(F)c(Cl)c4)c3cc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCC1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCN(Cc2ccc(Nc3nc4ncnc(Nc5ccc(OCc6cccc(F)c6)c(Cl)c5)c4s3)cc2)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)n1nc(-c2cccc(NS(C)(=O)=O)c2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1ccc(Nc2ncnc3c2NCc2cc(Br)ccc2O3)cc1Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(N(C)CCN(C)C)c(NC(=O)/C=C\C(C)C)cc1Nc1nc(C)cc(-c2cn(C)c3ccc(F)cc23)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(N2CCN(C)CC2)ccc1Nc1ncc(Cl)c(Oc2cccc(COc3no[n+]([O-])c3S(=O)(=O)c3ccccc3)c2)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CO[C@H]1[C@@H](N(C)C(=O)c2cccnc2)C[C@@H]2O[C@@]1(C)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[N+](C)(C/C=C/C(=O)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1)Cc1ccccc1[N+](=O)[O-].[Br-]>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(Br)c(/C=C2\CN(C)C/C(=C\c3nc(C)c(C)nc3C)C2=O)cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(Nc2ncnc3cc(OC(C)C)c4c(c23)OCCO4)cc1C#N>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC(=O)COCc1ccn2ncnc(Nc3ccc4c(cnn4Cc4cccc(F)c4)c3)c12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(-n2c(=O)cnc3cnc(Nc4ccc(N)cc4)nc32)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(C#CCO)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1ccncc1)Nc1cc(-c2cncnc2)cc(C(F)(F)F)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3cccc(Cl)c3F)c2cc1CN1CCN(C)CC1C(N)=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc2c(Nc3ccc(OCc4ccccn4)c(Cl)c3)ncnc2cc1O[C@H]1CCOC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)N1N=C(c2ccc(Cl)c(Cl)c2)CC1c1ccc(Cl)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N(C)c1nc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1OCCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CO[C@H]1[C@@H](N(C)CC(=O)O)C[C@@H]2O[C@@]1(C)n1c3ccccc3c3c4c(c5c6ccccc6n2c5c31)C(=O)NC4>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CC[C@H](N2C(=O)N(c3cccc(OC)c3F)Cc3cnc(Nc4ccc(N5CCN(C)CC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#CC(=O)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(N2C(=O)N(C)Cc3cnc(Nc4ccccc4)nc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncnc(Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)c1C#Cc1ccc(CNC(C)C)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C2CC(c3ccc(Cl)cc3)=NN2C2=NC(=O)CS2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3cccc(C#CC(C)(C)O)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(=O)Nc1cc2ncnc(NCc3ccccc3)c2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1CSC(N/N=C/c2cccc(F)c2)=N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#C/C(=C\c1cc(-c2ccccc2)c(O)c(-c2ccccc2)c1)C(N)=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=O)CCc1c(SSc2[nH]c3ccccc3c2CCC(N)=O)[nH]c2ccccc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=S(=O)(CCOCc1ccc(-c2ccc3ncnc(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)c3c2)o1)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(Nc2ncc3c(n2)N([C@H]2CCN(C(=O)C(C)(C)C)C2)C(=O)N(c2ccccc2Cl)C3)ccc1N1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(=O)c1cn2ncnc(Nc3ccc4[nH]ncc4c3)c2c1CC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cccc(Nc2ncnc3cc(NCCCN4CCN(C)CC4)ncc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1ccc2ncnc(Nc3cccc(Cl)c3)c2c1)c1cccc2c1OCCO2>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OC[C@@H](Nc1ncnc2sc3c(c12)CCCC3)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cl.O=C(CCN1CCOCC1)c1ccc(OCc2ccccc2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN(CC)CCC#CC(=O)Nc1ccc2ncnc(Nc3ccc(F)c(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@@H](Oc1cccc2ncnc(Nc3ccc4c(cnn4Cc4ccccn4)c3)c12)C(=O)N(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1cc2ncnc(Nc3cccc(Br)c3)c2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Br.Oc1ccc(-c2cc3c(NCc4ccccc4F)ncnc3[nH]2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/c1ccccc1)Nc1ccccc1F>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC/C=C\C(=O)Nc1cc(Nc2ncc(C#N)c(-c3cn(C)c4ccc(F)cc34)n2)c(OC)cc1N(C)CCN(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCc1ncnc(Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)c1C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1O[C@H]1CCOC1)/C(F)=C\CN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(-c2nc3cc(NC(=O)CCl)ccc3[nH]2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=OCCCCc1nc(-c2ccc(F)cc2)c(-c2ccnc3[nH]c(-c4ccccc4)cc23)[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C[C@@H](Nc1ncnc2oc(-c3ccccc3O)cc12)c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Brc1cccc(Nc2ncnc3cnc(NCCCn4ccnc4)cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(Nc2nc(Nc3ccc(/C=C/c4cc(C)cc(C)c4)cc3)ncc2F)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(=O)/C=C/C(=O)Cc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cnc(Nc2ccc(OCCN3CCCC3)cc2)nc1Nc1cccc(S(=O)(=O)NC(C)(C)C)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(CC(O)CO)c1cc2ncnc(Nc3cccc(Br)c3)c2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(Nc2ncc3c(n2)N([C@H]2CCN(C(=O)C(C)C)C2)C(=O)N(c2ccccc2Cl)C3)ccc1N1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCN1CC(O)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cl.NCCc1cn(CCCCCc2ccccc2)c2ccc(OCCc3ccc(O)cc3)cc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN(CC)c1ccc(Nc2cc3c(cc2Nc2ccc(N(CC)CC)cc2)C(=O)NC3=O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=S)N1N=C(c2ccc(Br)c(Br)c2)CC1c1ccccc1Br>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1Cc2c(cccc2Oc2ccc(Nc3ncnc4ccn(CCO)c34)cc2Cl)N1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCCCN(CCCC)C(=O)c1ccc(N(CCCl)CCCl)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCN(Cc2ccc(NC(=O)c3cccc(NC(=O)c4cnc5[nH]ccc5c4)c3)cc2C(F)(F)F)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(C#CCCN1CCOCC1)Nc1ccc2ncnc(Nc3ccc(F)c(Cl)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N1CC[C@H](N2C(=O)N(c3ccc(OC)cc3)Cc3cnc(Nc4ccc(N5CCN(C)CC5)c(C)c4)nc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCN(c1ccc(OC)cc1)c1nc(C)nc2oc(C)cc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC1(C)OB(c2ccc3ncnc(Nc4ccc(F)c(Cl)c4)c3c2)OC1(C)C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCCN1CCN(C(=O)CCCCCCCCCC(=O)N[C@H](C(=O)N2C[C@@H](O)C[C@@H]2C(=O)NCc2ccc(-c3scnc3C)cc2)C(C)(C)C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1C#CC(C)(C)N1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(Oc2nc(Nc3ccc(N4CCN(CC(=O)OCCCCCCOc5no[n+]([O-])c5S(=O)(=O)c5ccccc5)CC4)cc3OC)ncc2Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(NCc1ccccc1)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncnc(Nc2ccc(OCc3cccc(F)c3)c(Cl)c2)c1C#CCCNCc1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Clc1ccccc1-c1nnc(Cc2nc3ccccc3[nH]2)o1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C2=NN(c3nc(-c4ccccc4)cs3)C(c3ccc(Br)cc3)C2)cc1C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(N2C(=O)N(c3ccc(-c4ccccc4)cc3)Cc3cnc(Nc4ccc(N5CCN(C)CC5)cc4OC)nc32)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(Nc1ccccc1)NN1C(=O)c2c(c3c4cccc(O)c4n(C4OC(CO)C(O)C(O)C4O)c3c3[nH]c4c(O)cccc4c23)C1=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCCN1CCCCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NS(=O)(=O)c1ccc(NC(=S)Nc2ccc3ncnc(Nc4cccc(Br)c4)c3c2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Brc1cccc(Nc2ncnc3cc4[nH]nnc4cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC(=O)CN1CCC(n2cc(-c3cccc(O)c3)c3c(N)ncnc32)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=c1ccc2sc(-c3ccc(Nc4ncnc5[nH]cnc45)cc3)nc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc2[nH]c3nc(N)nc(NCc4cccc(Cl)c4)c3c2cc1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=S=C(Nc1ccc(Cl)c(Cl)c1)Nc1ccc2ncnc(Nc3cccc(Br)c3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3ccc(F)c(Cl)c3)c2cc1OCCO>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc2c1ncn2-c1ccccc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Brc1cccc(Nc2ncnc3ccc4cc[nH]c4c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncc(C#N)c(Nc3ccc(Cl)cc3F)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)N[C@H]1CC[C@H](N2C(=O)N(c3ccccc3Cl)Cc3cnc(Nc4ccc(N5CCN(C)CC5)c(C)c4)nc32)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)CCNc1cc2ncnc(Nc3cccc(Br)c3)c2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)c1ccc(/C=C/C(=O)c2ccc3ncc(C(N)=O)c(Nc4ccc(Cl)cc4)c3c2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1ccc2ncnc(Nc3ccc(C(C)(C)C)cc3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CCl)OCCn1c(=O)oc2cc3ncnc(Nc4ccc(N5CCCCC5)c(Cl)c4)c3cc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)CCNc1nc(-c2ccc3ncnc(Nc4ccc(OCc5cccc(C(F)(F)F)c5)cc4)c3c2)cs1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)CCCCC(=O)Nc1cccc(-c2c(-c3ccccc3)oc3ncnc(N[C@H](CO)c4ccccc4)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Nc1ncnc2c1c(-c1ccccc1)cn2C1CC(O)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC1CCN(Cc2ccn3ncnc(Nc4ccc(OCc5ccccn5)c(Cl)c4)c23)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)(C)NC(=O)c1cccc(Oc2ccc(Nc3ncnc4ccn(CCO)c34)cc2Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC(=O)C1=Cc2c(ncnc2Nc2ccc(Oc3cccc4sccc34)c(Cl)c2)NCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3ccc(Cl)cc3F)ncnc2cc1OCCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(-c2nccc(Nc3cc(C)[nH]n3)n2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCC(=O)Nc1cc2c(Nc3ccc4c(ccn4Cc4ccccn4)c3)ncnc2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(NC(=O)Nc2ccnc(Nc3ccc(OC)cc3OC)n2)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(CN1CCCC1)Nc1ccc2ncnc(Nc3ccc(Cl)cc3)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN1CCN(Cc2ccc(-c3ccc4ncnc(Nc5ccc(OCc6cccc(F)c6)c(Cl)c5)c4c3)o2)CC1.Cc1ccc(S(=O)(=O)O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C1COc2cc3ncnc(Nc4cccc(C(F)(F)F)c4)c3cc2N1CCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)C/C=C/C(=O)Nc1cccc(Nc2nc(N/N=C/c3ccc(F)cc3)ncc2Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3cc(OC(C)COC)c4c(c23)OCCO4)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1ccc(-n2c(=O)cnc3cnc(Nc4ccc(N5CCN(C)CC5)cc4)nc32)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#CC(C#N)=CNc1ccc(O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/c1ccccc1)Nc1cc(Cl)cc(Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncnc(Nc3cccc(Cl)c3F)c2cc1CN[C@H](C)C(N)=O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Brc1cc2c(NCc3cccs3)ncnc2s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(C2=C(c3c[nH]c4ccc(Br)cc34)C(=O)NC2=O)cc(OC)c1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOc1cc(/C=C(\C#N)C(N)=O)cc(CSc2ccc(C)cc2)c1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCOC(=O)C1=C(N)N(c2cccnc2)C2=C(C(=O)CCC2)C1c1cc2cc(Cl)ccc2n2nnnc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=NC(=S)NNc1nncc2ccccc12>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Brc1cccc(Nc2ncnc3ccncc23)c1NCCCCN1CCOCC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(C#CCCN1CCCCC1)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CS(=O)(=O)CCNC(=O)Nc1cc2c(Nc3ccc(F)c(Cl)c3)ncnc2cc1O[C@H]1CCOC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cc(Nc2nccc(NC(=O)c3cccc4ccccc34)n2)c(OC)cc1N1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=O=C(/C=C/CN1CCCCC1)N1CCOc2cc3ncnc(Nc4ccc(F)c(Cl)c4)c3cc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1ccc2ncnc(Nc3ccc(Br)cc3C)c2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Clc1cccc(CNc2[nH]nc3ncnc(Nc4cccc(Cl)c4)c23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc(C=C(C#N)C#N)ccc1O>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(C2=NN(c3nc(-c4ccc(Cl)cc4)cs3)C(c3ccc(C)cc3)C2)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#CCNCc1ccc(-c2cc3ncnc(Nc4ccc(OCc5cccc(F)c5)c(Cl)c4)c3s2)o1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CCC[C@@H](n2nc(-c3ccc(C(F)(F)F)cc3)c3c(N)ncnc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C#Cc1cccc(Nc2ncnc3cc(OC)c(NC(=O)/C=C/CNC4CC4)cc23)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C2=NN(C3=NC(=O)CS3)C(c3cccc4ccccc34)C2)cc1C>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ncc([N+](=O)[O-])n1C/C(=N/NC(=O)c1ccc(O)cc1)c1ccc(Br)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2ncc(C#N)c(Nc3cccc(O)c3)c2cc1OC>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#Cc1cnc2cc3c(cc2c1Nc1cccc(Br)c1)OCO3>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1cc(Nc2ncc3c(n2)N([C@H]2CCN(C(=O)CC(C)C)C2)C(=O)N(c2ccccc2Cl)C3)ccc1N1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1cc2c(Nc3c(F)cc(Br)cc3F)ncnc2cc1OCC1CCN(C)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CN(C)CCCNC(=O)/C=C/C(=O)Cc1cc2c(Nc3cccc(Br)c3)ncnc2cn1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cc1ccc(C(=O)Nc2ccc(CN3CCN(C)CC3)c(C(F)(F)F)c2)cc1NC(=O)c1cccnc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CCC[C@@H](n2nc(-c3ccc(N(C)C)c4ccccc34)c3c(N)ncnc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Clc1ccc(C2=NN(c3ccccc3)C(c3cccc4ccccc34)C2)cc1Cl>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N1CCC(Oc2nc(Nc3ccc(N4CCOCC4)cc3)nc3[nH]cc(-c4ccnc(NC)c4)c23)CC1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)N[C@@H]1CCC[C@@H](n2c(NC(=O)c3cccc(C(F)(F)F)c3)nc3cc(C)ccc32)C1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)Nc1cccc(Nc2nc(N/N=C(\C)c3ccc(C)cc3)ncc2Cl)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Fc1cccc(Nc2ncnc3cc(OCCCN4CCOCC4)c4c(c23)OCCO4)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=N#CC(C#N)=Cc1cc(O)c(O)c(O)c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc(NC(=O)/C=C/CN(C)C)cc1Nc1ncc(Cl)c(-c2cnn3ccccc23)n1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COc1ccc2c(Nc3cccc(Br)c3)ncnc2c1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=C=CC(=O)NCCSc1nc(-c2ccc(F)c(C(F)(F)F)c2)c(-c2ccnc(Nc3ccccc3)c2)[nH]1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=S=C(Nc1ccc2ncnc(Nc3cccc(Cl)c3)c2c1)Nc1ncc(Cl)s1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)n1nc(-c2ccc3c(ccn3C(=O)OC(C)(C)C)c2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=Cn1cnc2c(F)c(Nc3ccc(Br)cc3Cl)c(C(=O)NOCCO)cc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CC(C)n1nc(-c2cc3cc(C=O)ccc3s2)c2c(N)ncnc21>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=COC(=O)c1ccc(-c2ccc(-c3ccccc3)cc2O)cc1>)>,
<ProteinLigandComplex with 2 components (<KLIFSKinase name=P00533>, <Ligand name=CCO/N=C/c1c(N)ncnc1Nc1ccc2c(cnn2Cc2cccc(F)c2)c1>)>,
...]
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic bonds are: [<openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f81a718b1b0> >, <openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f81a718ba20> >]
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: True
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 16, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 23, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 25, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 31, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic bonds are: [<openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f81a7219e40> >, <openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f81a7219510> >, <openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f81a72194b0> >, <openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f81a7219d80> >]
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 38, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 55, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic bonds are: [<openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f81a71e6cc0> >]
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 16, name: , idx: 13, aromatic: False, chiral: True with bonds:
bond order: 2, chiral: False to atom atomic num: 7, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: True, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 44, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 37, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 59, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 35, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 54, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 65, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 52, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 59, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 71, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 29, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 16, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 24, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: True, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: True, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 23, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: False
Atom atomic num: 7, name: , idx: 27, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 16, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 48, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 16, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 15, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 0, aromatic: False, chiral: False
bond order: 2, chiral: False to atom atomic num: 8, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 27, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 44, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 16, name: , idx: 2, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 36, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 37, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 38, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 37, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 38, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 39, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 25, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
Atom atomic num: 7, name: , idx: 28, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: True
Atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 28, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 62, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 41, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 10, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 58, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 37, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 45, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 36, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 42, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 37, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 40, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 39, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 41, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 42, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 69, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 16, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 44, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 16, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 42, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 56, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 66, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 58, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: True
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 16, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 41, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 16, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 2, chiral: False to atom atomic num: 7, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 27, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 31, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic bonds are: [<openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f823cfe34e0> >]
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 50, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 29, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 25, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
Atom atomic num: 7, name: , idx: 29, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 60, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 67, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 20, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 43, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 39, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 47, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 18, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 47, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 28, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 24, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 16, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 52, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 40, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 20, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 16, name: , idx: 40, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 37, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 62, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 47, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 55, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic bonds are: [<openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f823cf485a0> >]
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 37, aromatic: False, chiral: False
Atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 10, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 15, aromatic: False, chiral: False
Problematic bonds are: [<openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f823cee94b0> >]
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 52, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 51, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 58, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 53, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 108, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 28, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 43, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 39, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 54, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 55, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 59, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 61, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 63, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 58, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: True
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 16, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 53, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 37, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 47, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 37, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 54, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 10, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 38, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 15, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 23, aromatic: False, chiral: False
bond order: 2, chiral: False to atom atomic num: 8, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 25, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 29, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 59, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 43, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 29, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 18, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: True, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 49, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 19, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 27, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: True, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 44, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 44, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 26, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 15, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 0, aromatic: False, chiral: False
bond order: 2, chiral: False to atom atomic num: 8, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 45, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 15, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 0, aromatic: False, chiral: False
bond order: 2, chiral: False to atom atomic num: 8, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 26, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 34, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 37, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 39, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 47, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 13, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 2, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 41, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 24, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 39, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 27, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 29, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 10, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 32, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 28, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 39, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 16, name: , idx: 31, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 33, aromatic: False, chiral: False
Atom atomic num: 16, name: , idx: 60, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 59, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 61, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 62, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 43, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 34, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 41, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Atom atomic num: 7, name: , idx: 38, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 37, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 39, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 40, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 37, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: True, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 31, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 28, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 27, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 10, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 37, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 16, name: , idx: 31, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 38, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 13, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 44, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 60, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 35, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 38, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 39, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: True, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 43, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 35, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 37, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 38, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 44, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 15, name: , idx: 26, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 27, aromatic: False, chiral: False
bond order: 2, chiral: False to atom atomic num: 8, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 29, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 72, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 29, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 39, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
Atom atomic num: 7, name: , idx: 35, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 37, aromatic: False, chiral: True
Atom atomic num: 6, name: , idx: 37, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 35, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 38, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 39, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 74, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 56, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 16, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 45, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 39, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 42, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 55, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 65, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 16, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 45, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 48, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 47, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 27, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 23, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Atom atomic num: 7, name: , idx: 12, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 54, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 28, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 10, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 21, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 38, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 54, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 39, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 43, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic bonds are: [<openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f823cc5bc00> >, <openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f823cc5bcc0> >]
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic bonds are: [<openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f823cbb9d20> >]
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 23, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: True, chiral: False
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 56, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic bonds are: [<openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f823ccc3b10> >]
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 52, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 34, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 31, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 34, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 34, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 38, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 29, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 36, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 37, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 38, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: True, chiral: False
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 38, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 40, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 10, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic bonds are: [<openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f823c85be70> >]
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 53, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 28, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 17, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 37, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 10, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 56, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
Atom atomic num: 7, name: , idx: 25, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 53, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: True, chiral: False
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 16, name: , idx: 12, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 54, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 52, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 49, aromatic: False, chiral: False
Atom atomic num: 7, name: , idx: 23, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 43, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 31, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 30, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 20, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 42, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 57, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 16, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 34, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 49, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 38, aromatic: False, chiral: False
Atom atomic num: 7, name: , idx: 6, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 51, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 45, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 59, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 27, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 60, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 30, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 64, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 32, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 66, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 68, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 1, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 0, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 24, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 45, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 18, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic bonds are: [<openeye.oechem.OEBondBase; proxy of <Swig Object of type 'OEChem::OEBondBase *' at 0x7f823cabdea0> >]
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 11, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 19, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 30, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 13, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 12, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 2, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 54, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 30, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 32, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 1, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 30, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 41, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 5, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 16, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 8, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 9, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 24, aromatic: False, chiral: True
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 54, aromatic: False, chiral: False
Atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 8, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 9, aromatic: False, chiral: True
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 16, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 17, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 18, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 7, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 25, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 10, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 32, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 31, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 41, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 33, aromatic: False, chiral: False
Atom atomic num: 6, name: , idx: 35, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 34, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 40, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 36, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 41, aromatic: False, chiral: False
Atom atomic num: 7, name: , idx: 38, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 37, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 39, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 40, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 14, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: True, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 33, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 27, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 26, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 29, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 8, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 36, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 7, name: , idx: 22, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 23, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 28, aromatic: False, chiral: True
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 5, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 13, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 38, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 3, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 7, name: , idx: 2, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 6, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 24, aromatic: False, chiral: False
Warning (not error because allow_undefined_stereo=True): OEMol has unspecified stereochemistry. oemol.GetTitle():
Problematic atoms are:
Atom atomic num: 6, name: , idx: 21, aromatic: False, chiral: True with bonds:
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 15, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 4, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 6, name: , idx: 22, aromatic: False, chiral: False
bond order: 1, chiral: False to atom atomic num: 1, name: , idx: 50, aromatic: False, chiral: False
Filter¶
Remove systems that couldn’t be featurized. Successful featurizations are stored in measurement.system.featurizations['last'] so we test for that key existence.
[12]:
from kinoml.datasets.groups import CallableGrouper, RandomGrouper
grouper = CallableGrouper(lambda measurement: 'invalid' if 'last' not in measurement.system.featurizations else 'valid')
grouper.assign(dataset, overwrite=True, progress=False)
groups = dataset.split_by_groups()
if "invalid" in groups:
_invalid = groups.pop("invalid")
warn(f"{len(_invalid)} entries could not be featurized!. Possible errors:")
warn(f"{_invalid[0].system.featurizations}")
Groups¶
Cumulatively apply groups.
[13]:
groups[("valid",)] = groups.pop("valid")
if GROUPS:
for grouper_str, grouper_kwargs in GROUPS:
grouper_cls = import_object(grouper_str)
## We need this because lambda functions are not JSON-serializable
if issubclass(grouper_cls, CallableGrouper):
for k, v in list(grouper_kwargs.items()):
if k == "function" and isinstance(v, str):
grouper_kwargs[k] = eval(v) # sorry :)
## End of lambda hack
grouper = grouper_cls(**grouper_kwargs)
for group_key in list(groups.keys()):
grouper.assign(groups[group_key], overwrite=True, progress=False)
for subkey, subgroup in groups.pop(group_key).split_by_groups().items():
groups[group_key + (subkey,)] = subgroup
print("10 groups to show keys:", *list(groups.keys())[:10], sep="\n")
10 groups to show keys:
('valid', 'pIC50Measurement')
('valid', 'pKdMeasurement')
('valid', 'pKiMeasurement')
Write tensors to disk¶
Output files are written to _output/<PIPELINE>/<DATASET>/<GROUP>.parquet files.
Each parquet will contain at least two array-like objects. The dimensionality of the parquet files is built as (systems, X_or_y, ...). For example, the first X vector for the first system is accessed like parquet[0, "0"]. Notice how the 2nd index is a string! (awkward design).
"0"(X, featurized systems). SeeDatasetProvider.to_awkwardfor more info."1"(y, associated measurements)
If X is composed of more than one array (e.g. connectivity matrix + node features), these are flattened to "0", "1", "2", and so on. y is ALWAYS the last one in that list (accessible via data.fields)
[14]:
random_grouper = RandomGrouper(TRAIN_TEST_VAL_KWARGS)
parquets = []
for group, ds in sorted(groups.items(), key=lambda kv: len(kv[1]), reverse=True):
indices = random_grouper.indices(ds)
X, y = ds.to_awkward()
parquet = ak.zip([*X, y], depth_limit=1)
path = OUT / f"{'__'.join([g for g in group if g != 'valid'])}.parquet"
parquets.append(path)
ak.to_parquet(parquet, path)
# TODO: Missing indices?
Preview generated Parquet files:
[15]:
from kinoml.datasets.torch_datasets import AwkwardArrayDataset
awk = AwkwardArrayDataset.from_parquet(parquets[0])
awk
[15]:
<Array [([0, ..., 0], 6.14), ..., ([...], ...)] type='2826 * (var * int64, ...'>
[16]:
# X, y = awk[0] # (multi-X) and y tensors for first system
[17]:
# X
[18]:
# y
[19]:
print("Run finished at", datetime.now())
Run finished at 2023-11-06 10:51:51.390702
Reproducibility logs¶
[20]:
# Free some memory first
del awk, parquets, groups, dataset
[21]:
from kinoml.utils import watermark
w = watermark()
Watermark
---------
Last updated: 2023-11-06T10:51:51.400767+01:00
Python implementation: CPython
Python version : 3.9.18
IPython version : 8.15.0
Compiler : GCC 12.3.0
OS : Linux
Release : 6.2.0-36-generic
Machine : x86_64
Processor : x86_64
CPU cores : 16
Architecture: 64bit
Hostname: aleph
Git hash: d2352d6e07d594ed9fd973b544529f89feb82d78
numpy : 1.26.0
sys : 3.9.18 | packaged by conda-forge | (main, Aug 30 2023, 03:49:32)
[GCC 12.3.0]
awkward: 2.4.6
Watermark: 2.4.3
nvidia-smi
----------
stdout:
Mon Nov 6 10:51:51 2023
+-----------------------------------------------------------------------------+
| NVIDIA-SMI 525.147.05 Driver Version: 525.147.05 CUDA Version: 12.0 |
|-------------------------------+----------------------+----------------------+
| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|===============================+======================+======================|
| 0 NVIDIA GeForce ... Off | 00000000:01:00.0 On | N/A |
| N/A 59C P8 20W / 80W | 49MiB / 16384MiB | 17% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
+-----------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=============================================================================|
| 0 N/A N/A 1260 G /usr/lib/xorg/Xorg 45MiB |
+-----------------------------------------------------------------------------+
conda info
----------
sys.version: 3.11.5 (main, Sep 11 2023, 13:54:46) [GC...
sys.prefix: /home/raquellrdc/anaconda3
sys.executable: /home/raquellrdc/anaconda3/bin/python
conda location: /home/raquellrdc/anaconda3/lib/python3.11/site-packages/conda
conda-build: /home/raquellrdc/anaconda3/bin/conda-build
conda-content-trust: /home/raquellrdc/anaconda3/bin/conda-content-trust
conda-convert: /home/raquellrdc/anaconda3/bin/conda-convert
conda-debug: /home/raquellrdc/anaconda3/bin/conda-debug
conda-develop: /home/raquellrdc/anaconda3/bin/conda-develop
conda-env: /home/raquellrdc/anaconda3/bin/conda-env
conda-index: /home/raquellrdc/anaconda3/bin/conda-index
conda-inspect: /home/raquellrdc/anaconda3/bin/conda-inspect
conda-metapackage: /home/raquellrdc/anaconda3/bin/conda-metapackage
conda-pack: /home/raquellrdc/anaconda3/bin/conda-pack
conda-render: /home/raquellrdc/anaconda3/bin/conda-render
conda-repo: /home/raquellrdc/anaconda3/bin/conda-repo
conda-server: /home/raquellrdc/anaconda3/bin/conda-server
conda-skeleton: /home/raquellrdc/anaconda3/bin/conda-skeleton
conda-token: /home/raquellrdc/anaconda3/bin/conda-token
conda-verify: /home/raquellrdc/anaconda3/bin/conda-verify
user site dirs: ~/.local/lib/python3.10
CIO_TEST: <not set>
CONDA_DEFAULT_ENV: kinoml_env
CONDA_EXE: /home/raquellrdc/anaconda3/bin/conda
CONDA_PREFIX: /home/raquellrdc/anaconda3/envs/kinoml_env
CONDA_PROMPT_MODIFIER: (kinoml_env)
CONDA_PYTHON_EXE: /home/raquellrdc/anaconda3/bin/python
CONDA_ROOT: /home/raquellrdc/anaconda3
CONDA_SHLVL: 1
CURL_CA_BUNDLE: <not set>
LD_PRELOAD: <not set>
PATH: /home/raquellrdc/anaconda3/envs/kinoml_env/bin:/home/raquellrdc/anaconda3/condabin:/home/raquellrdc/.local/bin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/usr/games:/usr/local/games:/snap/bin
PYTHONHASHSEED: 1234
REQUESTS_CA_BUNDLE: <not set>
SSL_CERT_FILE: <not set>
XDG_SEAT_PATH: /org/freedesktop/DisplayManager/Seat0
XDG_SESSION_PATH: /org/freedesktop/DisplayManager/Session1
conda list
----------
# packages in environment at /home/raquellrdc/anaconda3/envs/kinoml_env:
#
# Name Version Build Channel
_libgcc_mutex 0.1 conda_forge conda-forge
_openmp_mutex 4.5 2_gnu conda-forge
_py-xgboost-mutex 2.0 gpu_0 conda-forge
alabaster 0.7.13 pypi_0 pypi
ambertools 23.3 py39h7fa913e_4 conda-forge
amberutils 21.0 pypi_0 pypi
ansiwrap 0.8.4 py_0 conda-forge
anyio 3.5.0 py39h06a4308_0
appdirs 1.4.4 pyh9f0ad1d_0 conda-forge
argon2-cffi 21.3.0 pyhd3eb1b0_0
argon2-cffi-bindings 21.2.0 py39h7f8727e_0
arpack 3.8.0 nompi_h0baa96a_101 conda-forge
arrow 1.3.0 pyhd8ed1ab_0 conda-forge
asttokens 2.0.5 pyhd3eb1b0_0
astunparse 1.6.3 pyhd8ed1ab_0 conda-forge
async-lru 2.0.4 pyhd8ed1ab_0 conda-forge
attrs 23.1.0 py39h06a4308_0
awkward 2.4.6 pyhd8ed1ab_0 conda-forge
awkward-cpp 24 py39h7633fee_0 conda-forge
aws-c-auth 0.7.4 h1083cbe_2 conda-forge
aws-c-cal 0.6.2 h09139f6_2 conda-forge
aws-c-common 0.9.3 hd590300_0 conda-forge
aws-c-compression 0.2.17 h184a658_3 conda-forge
aws-c-event-stream 0.3.2 h6fea174_2 conda-forge
aws-c-http 0.7.13 hb59894b_2 conda-forge
aws-c-io 0.13.33 h161b759_0 conda-forge
aws-c-mqtt 0.9.7 h55cd26b_0 conda-forge
aws-c-s3 0.3.17 hfb4bb88_4 conda-forge
aws-c-sdkutils 0.1.12 h184a658_2 conda-forge
aws-checksums 0.1.17 h184a658_2 conda-forge
aws-crt-cpp 0.24.2 ha28989d_2 conda-forge
aws-sdk-cpp 1.11.156 h314d761_4 conda-forge
babel 2.11.0 py39h06a4308_0
backcall 0.2.0 pyhd3eb1b0_0
beautifulsoup4 4.12.2 py39h06a4308_0
biopandas 0.4.1 pyhd8ed1ab_1 conda-forge
biopython 1.77 py39h27cfd23_0
biotite 0.38.0 py39h44dd56e_0 conda-forge
black 23.3.0 py39h06a4308_0
blas 1.0 mkl
bleach 4.1.0 pyhd3eb1b0_0
blosc 1.21.5 h0f2a231_0 conda-forge
bokeh 3.2.1 py39h2f386ee_0
bravado 11.0.3 pyhd8ed1ab_0 conda-forge
bravado-core 5.17.1 pyhd8ed1ab_0 conda-forge
brotli 1.0.9 he6710b0_2
brotli-python 1.0.9 py39h6a678d5_7
bzip2 1.0.8 h7b6447c_0
c-ares 1.20.1 hd590300_0 conda-forge
c-blosc2 2.10.5 hb4ffafa_0 conda-forge
ca-certificates 2023.08.22 h06a4308_0
cached-property 1.5.2 hd8ed1ab_1 conda-forge
cached_property 1.5.2 pyha770c72_1 conda-forge
cachetools 5.3.1 pyhd8ed1ab_0 conda-forge
cairo 1.18.0 h3faef2a_0 conda-forge
certifi 2023.7.22 py39h06a4308_0
cffi 1.15.1 py39h5eee18b_3
cftime 1.6.2 py39h7deecbd_0
charset-normalizer 2.0.4 pyhd3eb1b0_0
click 8.0.4 py39h06a4308_0
cloudpickle 2.2.1 py39h06a4308_0
codecov 2.1.13 pyhd8ed1ab_0 conda-forge
colorama 0.4.6 py39h06a4308_0
comm 0.1.2 py39h06a4308_0
contourpy 1.0.5 py39hdb19cb5_0
coverage 7.2.2 py39h5eee18b_0
cpuonly 2.0 0 pytorch
cuda-version 11.8 h70ddcb2_2 conda-forge
cudatoolkit 11.8.0 h4ba93d1_12 conda-forge
cycler 0.11.0 pyhd3eb1b0_0
cytoolz 0.12.0 py39h5eee18b_0
dask 2023.10.0 pyhd8ed1ab_0 conda-forge
dask-core 2023.10.0 pyhd8ed1ab_0 conda-forge
dask-jobqueue 0.8.2 pyhd8ed1ab_0 conda-forge
debugpy 1.6.7 py39h6a678d5_0
decorator 5.1.1 pyhd3eb1b0_0
defusedxml 0.7.1 pyhd3eb1b0_0
distributed 2023.10.0 pyhd8ed1ab_0 conda-forge
docker-pycreds 0.4.0 pypi_0 pypi
docutils 0.20.1 pypi_0 pypi
edgembar 0.2 pypi_0 pypi
entrypoints 0.4 py39h06a4308_0
exceptiongroup 1.0.4 py39h06a4308_0
execnet 1.9.0 pyhd3eb1b0_0
executing 0.8.3 pyhd3eb1b0_0
expat 2.5.0 h6a678d5_0
fasteners 0.16.3 pyhd3eb1b0_0
ffmpeg 4.3 hf484d3e_0 pytorch
fftw 3.3.10 nompi_hc118613_108 conda-forge
filelock 3.9.0 py39h06a4308_0
font-ttf-dejavu-sans-mono 2.37 hd3eb1b0_0
font-ttf-inconsolata 2.001 hcb22688_0
font-ttf-source-code-pro 2.030 hd3eb1b0_0
font-ttf-ubuntu 0.83 h8b1ccd4_0
fontconfig 2.14.2 h14ed4e7_0 conda-forge
fonts-anaconda 1 h8fa9717_0
fonts-conda-ecosystem 1 hd3eb1b0_0
fonttools 4.25.0 pyhd3eb1b0_0
fqdn 1.5.1 pyhd8ed1ab_0 conda-forge
freetype 2.12.1 h4a9f257_0
fsspec 2023.9.2 py39h06a4308_0
gettext 0.21.1 h27087fc_0 conda-forge
gflags 2.2.2 he6710b0_0
giflib 5.2.1 h5eee18b_3
gitdb 4.0.11 pypi_0 pypi
gitpython 3.1.40 pypi_0 pypi
glog 0.6.0 h6f12383_0 conda-forge
gmp 6.2.1 h295c915_3
gmpy2 2.1.2 py39heeb90bb_0
gnutls 3.6.15 he1e5248_0
greenlet 2.0.1 py39h6a678d5_0
griddataformats 1.0.1 pyhd8ed1ab_0 conda-forge
gsd 3.2.0 py39h44dd56e_0 conda-forge
h5py 3.10.0 nompi_py39h87cadad_100 conda-forge
hdf4 4.2.15 h9772cbc_5 conda-forge
hdf5 1.14.2 nompi_h4f84152_100 conda-forge
heapdict 1.0.1 pyhd3eb1b0_0
icu 73.2 h59595ed_0 conda-forge
idna 3.4 py39h06a4308_0
imagesize 1.4.1 pypi_0 pypi
importlib-metadata 6.0.0 py39h06a4308_0
importlib-resources 6.1.0 pyhd8ed1ab_0 conda-forge
importlib_metadata 6.0.0 hd3eb1b0_0
importlib_resources 6.1.0 pyhd8ed1ab_0 conda-forge
iniconfig 1.1.1 pyhd3eb1b0_0
intel-openmp 2023.1.0 hdb19cb5_46305
ipykernel 6.25.0 py39h2f386ee_0
ipython 8.15.0 py39h06a4308_0
ipython_genutils 0.2.0 pyhd3eb1b0_1
ipywidgets 8.0.4 py39h06a4308_0
isoduration 20.11.0 pyhd8ed1ab_0 conda-forge
jedi 0.18.1 py39h06a4308_1
jinja2 3.1.2 py39h06a4308_0
joblib 1.2.0 py39h06a4308_0
jpeg 9e h5eee18b_1
json5 0.9.6 pyhd3eb1b0_0
jsonpointer 2.4 py39hf3d152e_3 conda-forge
jsonref 0.2 py_0 conda-forge
jsonschema 4.19.1 pyhd8ed1ab_0 conda-forge
jsonschema-specifications 2023.7.1 pyhd8ed1ab_0 conda-forge
jsonschema-with-format-nongpl 4.19.1 pyhd8ed1ab_0 conda-forge
jupyter-lsp 2.2.0 pyhd8ed1ab_0 conda-forge
jupyter_client 7.4.9 py39h06a4308_0
jupyter_core 5.3.0 py39h06a4308_0
jupyter_events 0.7.0 pyhd8ed1ab_2 conda-forge
jupyter_server 2.7.3 pyhd8ed1ab_1 conda-forge
jupyter_server_terminals 0.4.4 pyhd8ed1ab_1 conda-forge
jupyterlab 4.0.7 pyhd8ed1ab_0 conda-forge
jupyterlab_pygments 0.1.2 py_0
jupyterlab_server 2.25.0 pyhd8ed1ab_0 conda-forge
jupyterlab_widgets 3.0.9 pyhd8ed1ab_0 conda-forge
kinoml 0+unknown pypi_0 pypi
kiwisolver 1.4.4 py39h6a678d5_0
krb5 1.20.1 h143b758_1
lame 3.100 h7b6447c_0
lcms2 2.12 h3be6417_0
ld_impl_linux-64 2.38 h1181459_1
lerc 3.0 h295c915_0
libabseil 20230802.1 cxx17_h59595ed_0 conda-forge
libaec 1.1.2 h59595ed_1 conda-forge
libarrow 13.0.0 h4121bdd_9_cpu conda-forge
libblas 3.9.0 1_h86c2bf4_netlib conda-forge
libboost 1.82.0 h109eef0_2
libboost-python 1.82.0 py39hda80f44_6 conda-forge
libbrotlicommon 1.1.0 hd590300_1 conda-forge
libbrotlidec 1.1.0 hd590300_1 conda-forge
libbrotlienc 1.1.0 hd590300_1 conda-forge
libcrc32c 1.1.2 h6a678d5_0
libcurl 8.4.0 h251f7ec_0
libdeflate 1.17 h5eee18b_1
libedit 3.1.20221030 h5eee18b_0
libev 4.33 h7f8727e_1
libevent 2.1.12 hdbd6064_1
libffi 3.4.4 h6a678d5_0
libgcc-ng 13.2.0 h807b86a_2 conda-forge
libgfortran-ng 13.2.0 h69a702a_2 conda-forge
libgfortran5 13.2.0 ha4646dd_2 conda-forge
libglib 2.78.0 hebfc3b9_0 conda-forge
libgomp 13.2.0 h807b86a_2 conda-forge
libgoogle-cloud 2.12.0 h19a6dae_3 conda-forge
libgrpc 1.58.1 he06187c_2 conda-forge
libiconv 1.17 h166bdaf_0 conda-forge
libidn2 2.3.4 h5eee18b_0
libjpeg-turbo 2.0.0 h9bf148f_0 pytorch
liblapack 3.9.0 5_h92ddd45_netlib conda-forge
libnetcdf 4.9.2 nompi_h80fb2b6_112 conda-forge
libnghttp2 1.57.0 h2d74bed_0
libnsl 2.0.0 h5eee18b_0
libnuma 2.0.16 h0b41bf4_1 conda-forge
libpng 1.6.39 h5eee18b_0
libprotobuf 4.24.3 hf27288f_1 conda-forge
libre2-11 2023.06.02 h7a70373_0 conda-forge
libsodium 1.0.18 h7b6447c_0
libsqlite 3.43.2 h2797004_0 conda-forge
libssh2 1.10.0 hdbd6064_2
libstdcxx-ng 13.2.0 h7e041cc_2 conda-forge
libtasn1 4.19.0 h5eee18b_0
libthrift 0.19.0 hb90f79a_1 conda-forge
libtiff 4.5.1 h6a678d5_0
libunistring 0.9.10 h27cfd23_0
libutf8proc 2.8.0 h166bdaf_0 conda-forge
libuuid 2.38.1 h0b41bf4_0 conda-forge
libwebp 1.3.2 h11a3e52_0
libwebp-base 1.3.2 h5eee18b_0
libxcb 1.15 h7f8727e_0
libxgboost 1.7.6 cuda118h4159b1e_5 conda-forge
libxml2 2.11.5 h232c23b_1 conda-forge
libxslt 1.1.37 h0054252_1 conda-forge
libzip 1.10.1 h2629f0a_3 conda-forge
libzlib 1.2.13 hd590300_5 conda-forge
lightning-utilities 0.9.0 py39h06a4308_0
llvm-openmp 14.0.6 h9e868ea_0
locket 1.0.0 py39h06a4308_0
lxml 4.9.3 py39hed45dcc_1 conda-forge
lz4 4.3.2 py39h5eee18b_0
lz4-c 1.9.4 h6a678d5_0
lzo 2.10 h516909a_1000 conda-forge
markupsafe 2.1.1 py39h7f8727e_0
matplotlib-base 3.8.0 py39he9076e7_2 conda-forge
matplotlib-inline 0.1.6 py39h06a4308_0
mda-xdrlib 0.2.0 pyhd8ed1ab_0 conda-forge
mdanalysis 2.3.0 py39h4661b88_1 conda-forge
mdtraj 1.9.9 py39h031bd0f_0 conda-forge
mistune 0.8.4 py39h27cfd23_1000
mkl 2023.1.0 h213fc3f_46343
mkl-service 2.4.0 py39h5eee18b_1
mkl_fft 1.3.8 py39h5eee18b_0
mkl_random 1.2.4 py39hdb19cb5_0
mmpbsa-py 16.0 pypi_0 pypi
mmtf-python 1.1.3 pyhd8ed1ab_0 conda-forge
monotonic 1.5 py_0
mpc 1.1.0 h10f8cd9_1
mpfr 4.0.2 hb69a4c5_1
mpmath 1.3.0 py39h06a4308_0
mrcfile 1.4.3 pyhd8ed1ab_0 conda-forge
msgpack-python 1.0.3 py39hd09550d_0
munkres 1.1.4 py_0
mypy_extensions 1.0.0 py39h06a4308_0
nbclassic 0.5.5 py39h06a4308_0
nbclient 0.5.13 py39h06a4308_0
nbconvert 6.5.3 pyhd8ed1ab_0 conda-forge
nbconvert-core 6.5.3 pyhd8ed1ab_0 conda-forge
nbconvert-pandoc 6.5.3 pyhd8ed1ab_0 conda-forge
nbformat 5.9.2 py39h06a4308_0
nbval 0.10.0 pyhd8ed1ab_0 conda-forge
nccl 2.19.3.1 h6103f9b_0 conda-forge
ncurses 6.4 h6a678d5_0
nest-asyncio 1.5.6 py39h06a4308_0
netcdf-fortran 4.6.1 nompi_hacb5139_102 conda-forge
netcdf4 1.6.4 nompi_py39h4282601_103 conda-forge
nettle 3.7.3 hbbd107a_1
networkx 3.1 py39h06a4308_0
nglview 3.0.8 pyh1da8cd4_0 conda-forge
notebook 7.0.5 pyhd8ed1ab_0 conda-forge
notebook-shim 0.2.2 py39h06a4308_0
numexpr 2.7.3 py39hde0f152_1 conda-forge
numpy 1.26.0 py39h5f9d8c6_0
numpy-base 1.26.0 py39hb5e798b_0
ocl-icd 2.3.1 h7f98852_0 conda-forge
ocl-icd-system 1.0.0 1 conda-forge
opencadd 0.2.2 pyhd8ed1ab_0 conda-forge
openeye-toolkits 2023.1.1 py39_0 openeye
openff-amber-ff-ports 0.0.3 pyh6c4a22f_0 conda-forge
openff-forcefields 2023.08.0 pyh1a96a4e_0 conda-forge
openff-interchange 0.2.2 pyhd8ed1ab_0 conda-forge
openff-interchange-base 0.2.2 pyhd8ed1ab_0 conda-forge
openff-toolkit 0.11.2 pyhd8ed1ab_2 conda-forge
openff-toolkit-base 0.11.2 pyhd8ed1ab_2 conda-forge
openff-units 0.1.8 pyh1a96a4e_1 conda-forge
openff-utilities 0.1.10 pyhd8ed1ab_0 conda-forge
openh264 2.1.1 h4ff587b_0
openjpeg 2.4.0 h3ad879b_0
openmm 8.0.0 py39he81762f_3 conda-forge
openssl 3.1.3 hd590300_0 conda-forge
orc 1.9.0 h208142c_3 conda-forge
overrides 7.4.0 pyhd8ed1ab_0 conda-forge
packaging 23.1 py39h06a4308_0
packmol 20.010 h86c2bf4_0 conda-forge
packmol-memgen 2023.2.24 pypi_0 pypi
pandas 2.1.1 py39hddac248_1 conda-forge
pandoc 2.19.2 h32600fe_2 conda-forge
pandocfilters 1.5.0 pyhd3eb1b0_0
panedr 0.7.2 pyhd8ed1ab_0 conda-forge
papermill 2.2.2 pyhd8ed1ab_0 conda-forge
parmed 4.1.0 py39h227be39_0 conda-forge
parso 0.8.3 pyhd3eb1b0_0
partd 1.4.0 py39h06a4308_0
pathspec 0.10.3 py39h06a4308_0
pathtools 0.1.2 pypi_0 pypi
pbr 5.11.1 pyhd8ed1ab_0 conda-forge
pcre2 10.40 hc3806b6_0 conda-forge
pdb4amber 22.0 pypi_0 pypi
perl 5.32.1 4_hd590300_perl5 conda-forge
pexpect 4.8.0 pyhd3eb1b0_3
pickleshare 0.7.5 pyhd3eb1b0_1003
pillow 10.0.1 py39ha6cbd5a_0
pint 0.19.2 pyhd8ed1ab_0 conda-forge
pip 23.3 pyhd8ed1ab_0 conda-forge
pixman 0.42.2 h59595ed_0 conda-forge
pkgutil-resolve-name 1.3.10 pyhd8ed1ab_1 conda-forge
platformdirs 3.10.0 py39h06a4308_0
pluggy 1.0.0 py39h06a4308_1
prometheus_client 0.14.1 py39h06a4308_0
prompt-toolkit 3.0.36 py39h06a4308_0
protobuf 4.24.4 pypi_0 pypi
psutil 5.9.0 py39h5eee18b_0
ptyprocess 0.7.0 pyhd3eb1b0_2
pure_eval 0.2.2 pyhd3eb1b0_0
py-cpuinfo 9.0.0 pyhd8ed1ab_0 conda-forge
py-xgboost 1.7.6 cuda118py39h6e70402_5 conda-forge
pyarrow 13.0.0 py39h6925388_9_cpu conda-forge
pycairo 1.21.0 py39h287db57_0
pycparser 2.21 pyhd3eb1b0_0
pydantic 1.10.13 py39hd1e30aa_0 conda-forge
pyedr 0.7.2 pyhd8ed1ab_0 conda-forge
pyg 2.4.0 py39_torch_2.1.0_cpu pyg
pygments 2.15.1 py39h06a4308_1
pymsmt 22.0 pypi_0 pypi
pyparsing 3.0.9 py39h06a4308_0
pyrsistent 0.18.0 py39heee7806_0
pysocks 1.7.1 py39h06a4308_0
pytables 3.9.1 py39hfbd31a7_0 conda-forge
pytest 7.4.2 pyhd8ed1ab_0 conda-forge
pytest-cov 4.1.0 pyhd8ed1ab_0 conda-forge
pytest-xdist 3.3.1 pyhd8ed1ab_0 conda-forge
python 3.9.18 h0755675_0_cpython conda-forge
python-constraint 1.4.0 py_0 conda-forge
python-dateutil 2.8.2 pyhd3eb1b0_0
python-fastjsonschema 2.16.2 py39h06a4308_0
python-json-logger 2.0.7 pyhd8ed1ab_0 conda-forge
python-lmdb 1.4.1 py39h6a678d5_0
python-tzdata 2023.3 pyhd3eb1b0_0
python_abi 3.9 4_cp39 conda-forge
pytorch 2.1.0 py3.9_cpu_0 pytorch
pytorch-lightning 2.1.0 pyhd8ed1ab_0 conda-forge
pytorch-mutex 1.0 cpu pytorch
pytraj 2.0.6 pypi_0 pypi
pytz 2023.3.post1 py39h06a4308_0
pyyaml 6.0 py39h5eee18b_1
pyzmq 25.1.1 py39hb257651_1 conda-forge
rdkit 2023.09.1 py39hce5ca95_0 conda-forge
rdma-core 28.9 h59595ed_1 conda-forge
re2 2023.06.02 h2873b5e_0 conda-forge
readline 8.2 h5eee18b_0
referencing 0.30.2 pyhd8ed1ab_0 conda-forge
reportlab 3.5.67 py39hfdd840d_1
requests 2.31.0 pyhd8ed1ab_0 conda-forge
rfc3339-validator 0.1.4 pyhd8ed1ab_0 conda-forge
rfc3986-validator 0.1.1 pyh9f0ad1d_0 conda-forge
rpds-py 0.10.6 py39h9fdd4d6_0 conda-forge
ruamel.yaml 0.17.35 py39hd1e30aa_0 conda-forge
ruamel.yaml.clib 0.2.6 py39h5eee18b_1
s2n 1.3.54 h06160fa_0 conda-forge
sander 22.0 pypi_0 pypi
scikit-learn 1.3.0 py39h1128e8f_0
scipy 1.11.3 py39h5f9d8c6_0
seaborn 0.12.2 py39h06a4308_0
send2trash 1.8.2 pyh41d4057_0 conda-forge
sentry-sdk 1.32.0 pypi_0 pypi
setproctitle 1.3.3 pypi_0 pypi
setuptools 68.0.0 py39h06a4308_0
simplejson 3.17.6 py39h7f8727e_0
six 1.16.0 pyhd3eb1b0_1
sklearn-pytorch 0.1.0 pypi_0 pypi
smirnoff99frosst 1.1.0 pyh44b312d_0 conda-forge
smmap 5.0.1 pypi_0 pypi
snappy 1.1.10 h9fff704_0 conda-forge
sniffio 1.2.0 py39h06a4308_1
snowballstemmer 2.2.0 pypi_0 pypi
sortedcontainers 2.4.0 pyhd3eb1b0_0
soupsieve 2.5 py39h06a4308_0
sphinx 7.2.6 pypi_0 pypi
sphinxcontrib-applehelp 1.0.7 pypi_0 pypi
sphinxcontrib-devhelp 1.0.5 pypi_0 pypi
sphinxcontrib-htmlhelp 2.0.4 pypi_0 pypi
sphinxcontrib-jsmath 1.0.1 pypi_0 pypi
sphinxcontrib-qthelp 1.0.6 pypi_0 pypi
sphinxcontrib-serializinghtml 1.1.9 pypi_0 pypi
sqlalchemy 2.0.21 py39h5eee18b_0
stack_data 0.2.0 pyhd3eb1b0_0
swagger-spec-validator 3.0.3 pyhd8ed1ab_0 conda-forge
sympy 1.11.1 py39h06a4308_0
tbb 2021.8.0 hdb19cb5_0
tblib 1.7.0 pyhd3eb1b0_0
tenacity 8.2.2 py39h06a4308_0
terminado 0.17.1 py39h06a4308_0
textwrap3 0.9.2 py_0 conda-forge
threadpoolctl 2.2.0 pyh0d69192_0
tidynamics 1.1.2 pyhd8ed1ab_0 conda-forge
tinycss2 1.2.1 py39h06a4308_0
tk 8.6.13 h2797004_0 conda-forge
toml 0.10.2 pyhd3eb1b0_0
tomli 2.0.1 py39h06a4308_0
toolz 0.12.0 py39h06a4308_0
torchaudio 2.1.0 py39_cpu pytorch
torchmetrics 0.11.4 py39h2f386ee_1
torchvision 0.16.0 pypi_0 pypi
tornado 6.3.3 py39h5eee18b_0
tqdm 4.66.1 pyhd8ed1ab_0 conda-forge
traitlets 5.7.1 py39h06a4308_0
types-python-dateutil 2.8.19.14 pyhd8ed1ab_0 conda-forge
typing 3.10.0.0 py39h06a4308_0
typing-extensions 4.7.1 py39h06a4308_0
typing_extensions 4.7.1 py39h06a4308_0
typing_utils 0.1.0 pyhd8ed1ab_0 conda-forge
tzdata 2023c h04d1e81_0
ucx 1.15.0 h64cca9d_0 conda-forge
uri-template 1.3.0 pyhd8ed1ab_0 conda-forge
urllib3 2.0.3 py39h06a4308_0
wandb 0.15.12 pypi_0 pypi
watermark 2.4.3 pyhd8ed1ab_0 conda-forge
wcwidth 0.2.5 pyhd3eb1b0_0
webcolors 1.13 pyhd8ed1ab_0 conda-forge
webencodings 0.5.1 py39h06a4308_1
websocket-client 0.58.0 py39h06a4308_4
wheel 0.41.2 py39h06a4308_0
widgetsnbextension 4.0.5 py39h06a4308_0
xgboost 1.7.6 cuda118py39h6e70402_5 conda-forge
xmltodict 0.13.0 pyhd8ed1ab_0 conda-forge
xorg-kbproto 1.0.7 h7f98852_1002 conda-forge
xorg-libice 1.1.1 hd590300_0 conda-forge
xorg-libsm 1.2.4 h7391055_0 conda-forge
xorg-libx11 1.8.7 h8ee46fc_0 conda-forge
xorg-libxext 1.3.4 h0b41bf4_2 conda-forge
xorg-libxrender 0.9.11 hd590300_0 conda-forge
xorg-libxt 1.3.0 hd590300_1 conda-forge
xorg-renderproto 0.11.1 h7f98852_1002 conda-forge
xorg-xextproto 7.3.0 h0b41bf4_1003 conda-forge
xorg-xproto 7.0.31 h27cfd23_1007
xyzservices 2022.9.0 py39h06a4308_1
xz 5.4.2 h5eee18b_0
yaml 0.2.5 h7b6447c_0
zeromq 4.3.4 h2531618_0
zict 3.0.0 py39h06a4308_0
zipp 3.11.0 py39h06a4308_0
zlib 1.2.13 hd590300_5 conda-forge
zlib-ng 2.0.7 h0b41bf4_0 conda-forge
zstd 1.5.5 hc292b87_0
[22]:
%%capture cap --no-stderr
w = watermark()
[23]:
import json
with open(OUT/ "watermark.txt", "w") as f:
f.write(cap.stdout)
with open(OUT / "hparams.json", "w") as f:
json.dump(_hparams, f, default=str, indent=2)
2. 💻Run the experiment¶
Train a model with PyTorch¶
Now, we will train a single model using only PyTorch.
Tensors are loaded from Parquet files generated in the
features/pipeline. Each Parquet becomes a Torch Dataset sublass.Random splits are applied for train/test/(val).
It will train a single for model for a number of epochs across all datasets: epoch> dataloader> minibatch.
The loss is computed through the
loss_adaptermethod in each measurement_type.If validation is enabled, early stopping and LR schedulers are applied.
✏ Define hyper parameters¶
[24]:
# Parameters
PARQUET_LOADER_CLS = "kinoml.datasets.torch_datasets.AwkwardArrayDataset"
PARQUET_FILES = [
"ligand-only-morgan1024-EGFR/_output/ligand__MorganFingerprintFeaturizer_nbits=1024_radius=2/ChEMBLDatasetProvider/*.parquet"]
# Model -- specified with the full import path to the class object
## Machine learning model that will be trained. Pass it as importable string.
MODEL_CLS = "kinoml.ml.torch_models.NeuralNetworkRegression"
## Keyword arguments for the model initialization
MODEL_KWARGS = {"hidden_shape": 350} # input_shape is defined dynamically during training
# OPTIMIZER
## Optimizer class. Pass it as an importable string.
OPTIMIZER = "torch.optim.Adam"
## Keyword arguments for the optimizer
OPTIMIZER_KWARGS = {"lr": 0.001, "eps": 1e-7, "betas": [0.9, 0.999]}
# LOSS FUNCTION
## Loss function class. Pass it as an importable string.
LOSS = "torch.nn.MSELoss"
## Keyword arguments for the loss function, if applicable
LOSS_KWARGS = {}
# TRAINING
## Maximum number of epochs the training will run. In practice it might be less due to early stopping
MAX_EPOCHS = 50
## Enable real-time validation: this will split the test set into two halves: test and validation.
## It will also enable LR scheduling and early stopping, based on the validation loss.
VALIDATION = True
## Options for the builtin early stopper (kinoml.ml.torch_loops.EarlyStopping)
EARLY_STOPPING_KWARGS = {}
# DATALOADER
DATALOADER_CLS = "torch.utils.data.DataLoader" # you can also use torch_geometric.data.DataLoader
## Minibatch size
BATCH_SIZE = 64
## Proportion of the dataset that will be split into a test set. If VALIDATION=True,
## this will also cover the validation set. So, 0.2 will mean: 0.8 training, 0.1 test, 0.1 valid.
TRAIN_TEST_SPLIT = 0.2
## Whether to shuffle the indices before splitting
SHUFFLE_SPLITS = True
## Read https://pytorch.org/docs/stable/data.html#dataloader-collate-fn
## IMPORTANT: This will be needed if your X tensors have different shapes across systems!
COLLATE_FN = None
# Plot bootstrapping
## Bootstrapping iterations for the performance plots
N_BOOTSTRAPS = 1
## Proportion of the data that is sampled in each iteration
BOOTSTRAP_SAMPLE_RATIO = 1
# Output
## Enable some extra output, like plots and logging statements.
VERBOSE = False
HERE = _dh[-1] #current path
⚠ From here on, you should not need to modify anything else to apply it to your own dataset 🤞
Define key paths for data and outputs:
[25]:
from pathlib import Path
from datetime import datetime
HERE = Path(HERE)
for parent in HERE.parents:
if next(parent.glob(".github/"), None):
REPO = parent
break
FEATURES_STORE = REPO /"examples"/"experiments"
OUT = HERE / "_output" / datetime.now().strftime("%Y%m%d-%H%M%S")
OUT.mkdir(parents=True, exist_ok=True)
print(f"This notebook: HERE = {HERE}")
print(f"This repo: REPO = {REPO}")
print(f"Features: FEATURES_STORE = {FEATURES_STORE}")
print(f"Outputs in: OUT = {OUT}")
This notebook: HERE = /home/raquellrdc/Desktop/postdoc/kinoml_tech_paper_final/kinoml/examples/experiments/ligand-only-morgan1024-EGFR
This repo: REPO = /home/raquellrdc/Desktop/postdoc/kinoml_tech_paper_final/kinoml
Features: FEATURES_STORE = /home/raquellrdc/Desktop/postdoc/kinoml_tech_paper_final/kinoml/examples/experiments
Outputs in: OUT = /home/raquellrdc/Desktop/postdoc/kinoml_tech_paper_final/kinoml/examples/experiments/ligand-only-morgan1024-EGFR/_output/20231106-105158
[26]:
# Nasty trick: save all-caps local variables (CONSTANTS working as hyperparameters) so far in a dict to save it later
_hparams = {key: value for key, value in locals().items() if key.upper() == key and not key.startswith(("_", "OE_"))}
[27]:
from collections import defaultdict
from warnings import warn
import sys
import shutil
from IPython.display import Markdown
import numpy as np
import pandas as pd
import torch
from torch.utils.data import DataLoader, SubsetRandomSampler
from tqdm.auto import trange, tqdm
from kinoml.ml.torch_loops import LRScheduler, EarlyStopping
from kinoml.utils import seed_everything, import_object
from kinoml.core import measurements as measurement_types
from kinoml.core.measurements import null_observation_model
from kinoml.analysis.metrics import performance
from kinoml.analysis.plots import predicted_vs_observed
# Fix the seed for reproducible random splits -- otherwise we get mixed train/test groups every time, biasing the model evaluation
seed_everything();
print("Run started at", datetime.now())
Run started at 2023-11-06 10:51:59.212444
Load featurized data and create observation models¶
We assume this path structure: $REPO/features/_output/<FEATURIZATION>/<DATASET>/<MEASUREMENT_TYPE>.npz
[28]:
DATASETS = []
MEASUREMENT_TYPES = set()
ParquetLoaderCls = import_object(PARQUET_LOADER_CLS)
for glob in PARQUET_FILES:
print(glob)
parquets = list(FEATURES_STORE.glob(glob))
if not parquets:
warn(f"⚠ Parquet glob `{glob}` did not match any files!")
continue
for parquet in parquets:
measurement_type = parquet.stem
dataset = parquet.parent.name
ds = ParquetLoaderCls.from_parquet(parquet)
ds.metadata = {
"dataset": dataset,
"measurement_type": measurement_type,
}
DATASETS.append(ds)
MEASUREMENT_TYPES.add(measurement_type)
if not DATASETS:
raise ValueError("Provided `PARQUET_FILES` did not result in any valid datasets!")
ligand-only-morgan1024-EGFR/_output/ligand__MorganFingerprintFeaturizer_nbits=1024_radius=2/ChEMBLDatasetProvider/*.parquet
Now that we have all the data-dependent objects, we can start with the model-specific definitions.
[29]:
print(f"Loaded {len(DATASETS)} datasets with a total of {sum(len(d) for d in DATASETS)} measurements.")
Loaded 3 datasets with a total of 3000 measurements.
Prepare splits and dataloaders¶
Create train / test / validation subsets. Here we implement a random split, but it can take external indices if needed.
[30]:
dataloaders = {}
for dataset in DATASETS:
key = dataset.metadata["measurement_type"]
# Generate random indices in situ
# If you need to provide indices from another source,
# replace this block to provide train_indices, test_indices
dataset_size = len(dataset)
indices = list(range(dataset_size))
split = int(np.floor(TRAIN_TEST_SPLIT * dataset_size))
if SHUFFLE_SPLITS :
np.random.shuffle(indices)
train_indices, test_indices = indices[split:], indices[split:]
if VALIDATION:
split2 = int(np.floor(len(test_indices) / 2))
test_indices, val_indices = test_indices[:split2], test_indices[split2:]
# End of indices creation
collate_fn = None
if COLLATE_FN:
# IMPORTANT: This will be needed if your X tensors have different shapes across systems!
# COLLATE_FN can be an import string, or a eval-able lambda
# Read https://pytorch.org/docs/stable/data.html#dataloader-collate-fn
try:
collate_fn = import_object(COLLATE_FN)
except ImportError:
collate_fn = eval(COLLATE_FN)
# Creating PT data samplers and loaders:
train_sampler = SubsetRandomSampler(train_indices)
test_sampler = SubsetRandomSampler(test_indices)
dataloaders[key] = {
"train": import_object(DATALOADER_CLS)(dataset, batch_size=BATCH_SIZE, sampler=train_sampler, collate_fn=collate_fn),
"test": import_object(DATALOADER_CLS)(dataset, batch_size=BATCH_SIZE, sampler=test_sampler, collate_fn=collate_fn),
}
if VALIDATION:
val_sampler = SubsetRandomSampler(val_indices)
dataloaders[key]["val"] = import_object(DATALOADER_CLS)(dataset, batch_size=BATCH_SIZE, sampler=val_sampler, collate_fn=collate_fn)
Training loop¶
[31]:
ModelCls = import_object(MODEL_CLS)
# Note that we assume all dataloaders will provide the
# same kind of input shape, so we onlt test on one
if ModelCls.needs_input_shape:
a_dataloader = dataloaders[next(iter(dataloaders.keys()))]["train"]
x_sample, _ = next(iter(a_dataloader))
MODEL_KWARGS["input_shape"] = ModelCls.estimate_input_shape(x_sample)
nn_model = ModelCls(**MODEL_KWARGS)
optimizer = import_object(OPTIMIZER)(nn_model.parameters(), **OPTIMIZER_KWARGS)
loss_function = import_object(LOSS)()
if VALIDATION:
lr_scheduler = LRScheduler(optimizer)
early_stopping = EarlyStopping(**EARLY_STOPPING_KWARGS)
train_loss_timeseries = []
val_loss_timeseries = []
range_epochs = trange(MAX_EPOCHS, desc="Epochs")
for epoch in range_epochs:
train_loss = 0.0
val_loss = 0.0
for key, loader in tqdm(dataloaders.items(), desc="Datasets", leave=False):
try:
mtype_class = import_object(f"kinoml.core.measurements.{key}")
except AttributeError:
mtype_class = import_object(f"kinoml.core.measurements.{key.split('__')[1]}")
loss_adapter = mtype_class.loss_adapter(backend="pytorch")
# TRAIN
nn_model.train()
for x, y in tqdm(loader["train"], desc="Minibatches", leave=False):
# Clear gradients
optimizer.zero_grad()
# Obtain model prediction given model input
prediction = nn_model(x)
# apply observation model
loss = loss_adapter(prediction.view_as(y), y, loss_function)
# Pred. must match y shape! ^^^^^^^^^^
# Obtain loss for the predicted output
train_loss += loss.item()
# Gradients w.r.t. parameters
loss.backward()
# Optimize
optimizer.step()
# VALIDATE
if VALIDATION:
nn_model.eval()
with torch.no_grad():
for x, y in tqdm(loader["val"], desc="Minibatches", leave=False):
prediction = nn_model(x).view_as(y)
loss = loss_adapter(prediction.view_as(y), y, loss_function)
val_loss += loss.item()
range_epochs.set_description(f"Epochs (Avg. val. loss={val_loss / (epoch + 1):.2e})")
# LOG LOSSES
train_loss_timeseries.append(train_loss)
if VALIDATION:
val_loss_timeseries.append(val_loss)
# Adjust training if needed
lr_scheduler(val_loss)
early_stopping(val_loss)
if early_stopping.early_stop:
break
INFO: Early stopping counter 1 of 5
INFO: Early stopping counter 2 of 5
INFO: Early stopping counter 3 of 5
Save model to disk
[32]:
torch.save(nn_model, OUT / "nn_model.pt")
Evaluate model¶
[33]:
metrics = {}
nn_model.train(False)
for key, loader in dataloaders.items():
metrics[key] = {}
display(Markdown(f"#### {key}"))
for ttype, dataloader in loader.items():
display(Markdown(f"##### {ttype}"))
try:
mtype= import_object(f"kinoml.core.measurements.{key}")
except AttributeError:
mtype = import_object(f"kinoml.core.measurements.{key.split('__')[1]}")
obs_model = mtype.observation_model(backend="pytorch")
x, y = dataloader.dataset[dataloader.batch_sampler.sampler.indices]
prediction = obs_model(nn_model(x).view_as(y).detach().numpy())
perf_data = performance(prediction, y, verbose=False)
metrics[key][ttype] = {}
for perfkey, values in perf_data.items():
metrics[key][ttype][perfkey] = {"mean": values[0], "std": values[1]}
display(predicted_vs_observed(prediction, y, mtype))
pKdMeasurement¶
train¶
MAE: 0.2498±0.0533 95CI=(0.1692, 0.3305)
MSE: 0.1704±0.0794 95CI=(0.0651, 0.3030)
R2: 0.8635±0.0715 95CI=(0.7471, 0.9571)
RMSE: 0.4013±0.0969 95CI=(0.2551, 0.5504)
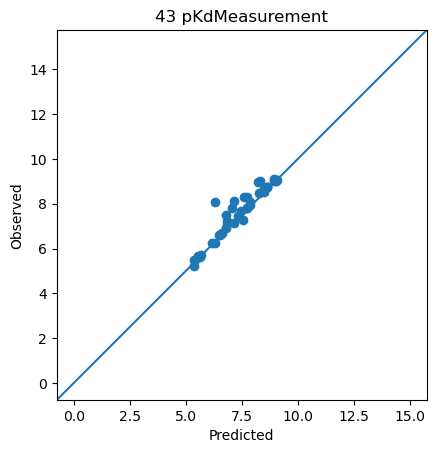
test¶
MAE: 0.2738±0.0872 95CI=(0.1629, 0.4197)
MSE: 0.2104±0.1424 95CI=(0.0542, 0.4710)
R2: 0.7849±0.2243 95CI=(0.4298, 0.9582)
RMSE: 0.4323±0.1535 95CI=(0.2328, 0.6863)
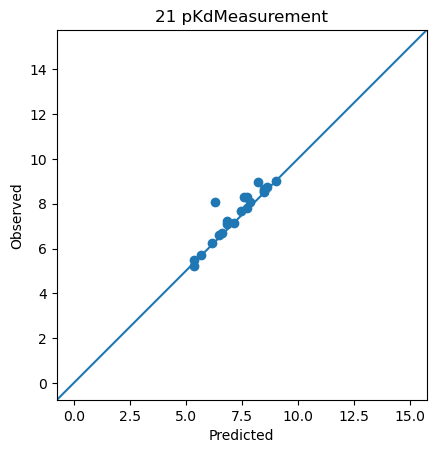
val¶
MAE: 0.2216±0.0618 95CI=(0.1283, 0.3213)
MSE: 0.1199±0.0543 95CI=(0.0401, 0.2115)
R2: 0.8932±0.0709 95CI=(0.7751, 0.9764)
RMSE: 0.3358±0.0849 95CI=(0.2002, 0.4599)
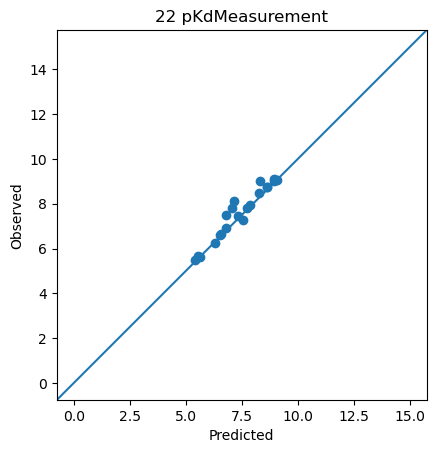
pIC50Measurement¶
train¶
MAE: 0.2354±0.0052 95CI=(0.2264, 0.2442)
MSE: 0.1034±0.0058 95CI=(0.0939, 0.1138)
R2: 0.9486±0.0028 95CI=(0.9436, 0.9527)
RMSE: 0.3214±0.0089 95CI=(0.3065, 0.3373)
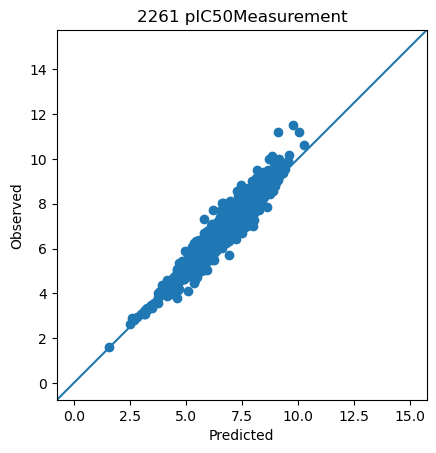
test¶
MAE: 0.2321±0.0073 95CI=(0.2209, 0.2425)
MSE: 0.1017±0.0077 95CI=(0.0898, 0.1157)
R2: 0.9498±0.0041 95CI=(0.9416, 0.9555)
RMSE: 0.3186±0.0120 95CI=(0.2997, 0.3402)
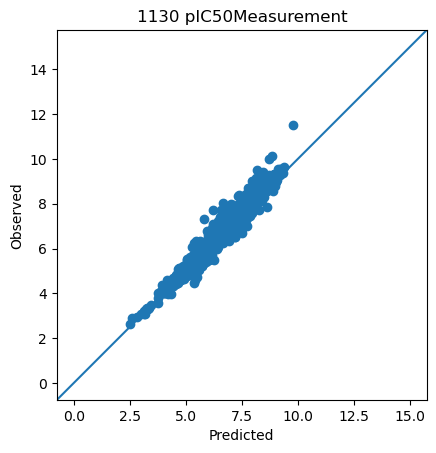
val¶
MAE: 0.2386±0.0071 95CI=(0.2269, 0.2499)
MSE: 0.1052±0.0082 95CI=(0.0934, 0.1183)
R2: 0.9472±0.0038 95CI=(0.9413, 0.9527)
RMSE: 0.3241±0.0126 95CI=(0.3056, 0.3439)
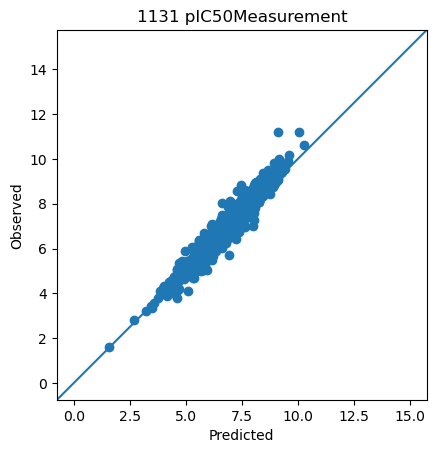
pKiMeasurement¶
train¶
MAE: 0.1778±0.0158 95CI=(0.1512, 0.2068)
MSE: 0.0529±0.0088 95CI=(0.0388, 0.0671)
R2: 0.9676±0.0079 95CI=(0.9546, 0.9781)
RMSE: 0.2292±0.0191 95CI=(0.1970, 0.2590)
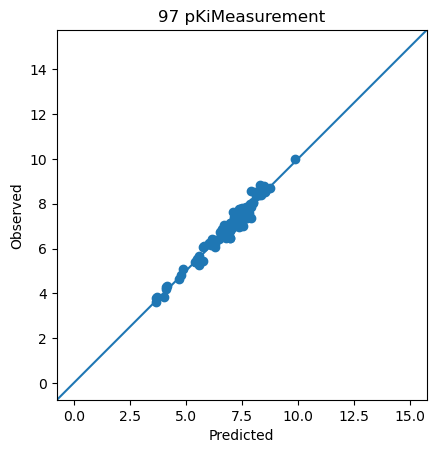
test¶
MAE: 0.1665±0.0186 95CI=(0.1355, 0.1965)
MSE: 0.0416±0.0085 95CI=(0.0278, 0.0576)
R2: 0.9811±0.0058 95CI=(0.9701, 0.9890)
RMSE: 0.2028±0.0207 95CI=(0.1668, 0.2399)
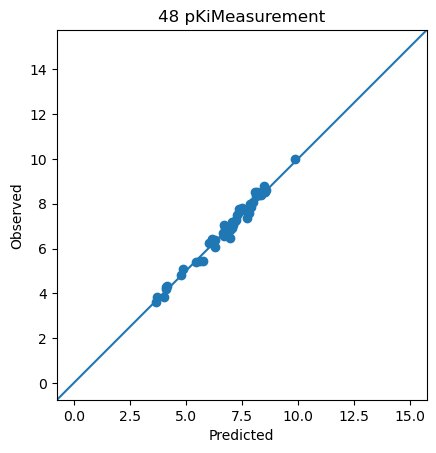
val¶
MAE: 0.1933±0.0262 95CI=(0.1510, 0.2438)
MSE: 0.0653±0.0154 95CI=(0.0398, 0.0926)
R2: 0.9303±0.0255 95CI=(0.8826, 0.9639)
RMSE: 0.2538±0.0303 95CI=(0.1995, 0.3043)
Summary¶
kinase_metrics is a nested dictionary with these dimensions:
measurement type
metric
mean & standard deviation
[34]:
import json
display(Markdown(f"""
### Configuration
```json
{json.dumps(_hparams, default=str, indent=2)}
```
"""))
if VERBOSE:
display(Markdown(
f"""
### Kinase metrics
```json
{json.dumps(metrics, default=str, indent=2)}
```
"""))
Configuration¶
{
"VERSION": "2023.11.06",
"HERE": "/home/raquellrdc/Desktop/postdoc/kinoml_tech_paper_final/kinoml/examples/experiments/ligand-only-morgan1024-EGFR",
"DATASET_CLS": "kinoml.datasets.chembl.ChEMBLDatasetProvider",
"DATASET_KWARGS": {
"path_or_url": "../data/filtered_dataset_EGFR.csv",
"measurement_types": [
"pIC50",
"pKd",
"pKi"
],
"uniprot_ids": [
"P00533"
],
"sample": 3000
},
"PIPELINES": {
"ligand": [
[
"kinoml.features.ligand.MorganFingerprintFeaturizer",
{
"nbits": 1024,
"radius": 2
}
]
]
},
"PIPELINES_AGG": "kinoml.features.core.TupleOfArrays",
"PIPELINES_AGG_KWARGS": {},
"FEATURIZE_KWARGS": {
"keep": false
},
"GROUPS": [
[
"kinoml.datasets.groups.CallableGrouper",
{
"function": "<function <lambda> at 0x7fce6846ea60>"
}
]
],
"TRAIN_TEST_VAL_KWARGS": {
"idx_train": 0.8,
"idx_test": 0.1,
"idx_val": 0.1
},
"REPO": "/home/raquellrdc/Desktop/postdoc/kinoml_tech_paper_final/kinoml",
"OUT": "/home/raquellrdc/Desktop/postdoc/kinoml_tech_paper_final/kinoml/examples/experiments/ligand-only-morgan1024-EGFR/_output/20231106-105158",
"X": "[[[0, 0, 0, 0, 0, 0, 0, 0, 1, 0, 0, ..., 0, 0, 0, 0, 0, 0, 1, 0, 0, 0, 0], ...]]",
"PARQUET_LOADER_CLS": "kinoml.datasets.torch_datasets.AwkwardArrayDataset",
"PARQUET_FILES": [
"ligand-only-morgan1024-EGFR/_output/ligand__MorganFingerprintFeaturizer_nbits=1024_radius=2/ChEMBLDatasetProvider/*.parquet"
],
"MODEL_CLS": "kinoml.ml.torch_models.NeuralNetworkRegression",
"MODEL_KWARGS": {
"hidden_shape": 350,
"input_shape": 1024
},
"OPTIMIZER": "torch.optim.Adam",
"OPTIMIZER_KWARGS": {
"lr": 0.001,
"eps": 1e-07,
"betas": [
0.9,
0.999
]
},
"LOSS": "torch.nn.MSELoss",
"LOSS_KWARGS": {},
"MAX_EPOCHS": 50,
"VALIDATION": true,
"EARLY_STOPPING_KWARGS": {},
"DATALOADER_CLS": "torch.utils.data.DataLoader",
"BATCH_SIZE": 64,
"TRAIN_TEST_SPLIT": 0.2,
"SHUFFLE_SPLITS": true,
"COLLATE_FN": null,
"N_BOOTSTRAPS": 1,
"BOOTSTRAP_SAMPLE_RATIO": 1,
"VERBOSE": false,
"FEATURES_STORE": "/home/raquellrdc/Desktop/postdoc/kinoml_tech_paper_final/kinoml/examples/experiments"
}
[35]:
for mtype_name in MEASUREMENT_TYPES:
mtype_metrics = metrics.get(mtype_name)
if not mtype_metrics:
continue
display(Markdown(f"#### {mtype_name}"))
# flatten dict a bit: from dict["test"]["r2"]["mean"] to dict["test"]["r2_mean"]
flattened = {}
for ttype, scores in mtype_metrics.items():
flattened[ttype] = {}
for score, stats in scores.items():
for stat, value in stats.items():
flattened[ttype][f"{score}_{stat}"] = value
df = pd.DataFrame.from_dict(flattened, orient="index")
with pd.option_context("display.float_format", "{:.3f}".format, "display.max_rows", len(df)):
display(
df.style.background_gradient(subset=["r2_mean"], low=0, high=1, vmin=0, vmax=1)
.apply(lambda x: ['font-weight: bold' for v in x], subset=["r2_mean"])
)
pKiMeasurement¶
| mae_mean | mae_std | mse_mean | mse_std | r2_mean | r2_std | rmse_mean | rmse_std | |
|---|---|---|---|---|---|---|---|---|
| train | 0.177771 | 0.015785 | 0.052921 | 0.008807 | 0.967586 | 0.007924 | 0.229248 | 0.019131 |
| test | 0.166451 | 0.018611 | 0.041571 | 0.008508 | 0.981131 | 0.005752 | 0.202837 | 0.020681 |
| val | 0.193261 | 0.026235 | 0.065315 | 0.015376 | 0.930327 | 0.025522 | 0.253769 | 0.030277 |
pIC50Measurement¶
| mae_mean | mae_std | mse_mean | mse_std | r2_mean | r2_std | rmse_mean | rmse_std | |
|---|---|---|---|---|---|---|---|---|
| train | 0.235393 | 0.005219 | 0.103362 | 0.005762 | 0.948592 | 0.002752 | 0.321376 | 0.008907 |
| test | 0.232105 | 0.007312 | 0.101654 | 0.007653 | 0.949771 | 0.004129 | 0.318607 | 0.011975 |
| val | 0.238604 | 0.007122 | 0.105168 | 0.008246 | 0.947172 | 0.003796 | 0.324051 | 0.012595 |
pKdMeasurement¶
| mae_mean | mae_std | mse_mean | mse_std | r2_mean | r2_std | rmse_mean | rmse_std | |
|---|---|---|---|---|---|---|---|---|
| train | 0.249806 | 0.053278 | 0.170412 | 0.079378 | 0.863479 | 0.071541 | 0.401279 | 0.096887 |
| test | 0.273753 | 0.087242 | 0.210392 | 0.142430 | 0.784935 | 0.224324 | 0.432251 | 0.153464 |
| val | 0.221559 | 0.061849 | 0.119945 | 0.054289 | 0.893225 | 0.070891 | 0.335753 | 0.084941 |
[36]:
print("Run finished at", datetime.now())
Run finished at 2023-11-06 10:53:44.645164
Save reports to disk¶
[37]:
from kinoml.utils import watermark
w = watermark()
Watermark
---------
Last updated: 2023-11-06T10:53:44.649391+01:00
Python implementation: CPython
Python version : 3.9.18
IPython version : 8.15.0
Compiler : GCC 12.3.0
OS : Linux
Release : 6.2.0-36-generic
Machine : x86_64
Processor : x86_64
CPU cores : 16
Architecture: 64bit
Hostname: aleph
Git hash: d2352d6e07d594ed9fd973b544529f89feb82d78
numpy : 1.26.0
json : 2.0.9
sys : 3.9.18 | packaged by conda-forge | (main, Aug 30 2023, 03:49:32)
[GCC 12.3.0]
kinoml : 0+unknown
torch : 2.1.0
awkward: 2.4.6
pandas : 2.1.1
Watermark: 2.4.3
nvidia-smi
----------
stdout:
Mon Nov 6 10:53:44 2023
+-----------------------------------------------------------------------------+
| NVIDIA-SMI 525.147.05 Driver Version: 525.147.05 CUDA Version: 12.0 |
|-------------------------------+----------------------+----------------------+
| GPU Name Persistence-M| Bus-Id Disp.A | Volatile Uncorr. ECC |
| Fan Temp Perf Pwr:Usage/Cap| Memory-Usage | GPU-Util Compute M. |
| | | MIG M. |
|===============================+======================+======================|
| 0 NVIDIA GeForce ... Off | 00000000:01:00.0 On | N/A |
| N/A 62C P8 21W / 80W | 49MiB / 16384MiB | 12% Default |
| | | N/A |
+-------------------------------+----------------------+----------------------+
+-----------------------------------------------------------------------------+
| Processes: |
| GPU GI CI PID Type Process name GPU Memory |
| ID ID Usage |
|=============================================================================|
| 0 N/A N/A 1260 G /usr/lib/xorg/Xorg 45MiB |
+-----------------------------------------------------------------------------+
conda info
----------
sys.version: 3.11.5 (main, Sep 11 2023, 13:54:46) [GC...
sys.prefix: /home/raquellrdc/anaconda3
sys.executable: /home/raquellrdc/anaconda3/bin/python
conda location: /home/raquellrdc/anaconda3/lib/python3.11/site-packages/conda
conda-build: /home/raquellrdc/anaconda3/bin/conda-build
conda-content-trust: /home/raquellrdc/anaconda3/bin/conda-content-trust
conda-convert: /home/raquellrdc/anaconda3/bin/conda-convert
conda-debug: /home/raquellrdc/anaconda3/bin/conda-debug
conda-develop: /home/raquellrdc/anaconda3/bin/conda-develop
conda-env: /home/raquellrdc/anaconda3/bin/conda-env
conda-index: /home/raquellrdc/anaconda3/bin/conda-index
conda-inspect: /home/raquellrdc/anaconda3/bin/conda-inspect
conda-metapackage: /home/raquellrdc/anaconda3/bin/conda-metapackage
conda-pack: /home/raquellrdc/anaconda3/bin/conda-pack
conda-render: /home/raquellrdc/anaconda3/bin/conda-render
conda-repo: /home/raquellrdc/anaconda3/bin/conda-repo
conda-server: /home/raquellrdc/anaconda3/bin/conda-server
conda-skeleton: /home/raquellrdc/anaconda3/bin/conda-skeleton
conda-token: /home/raquellrdc/anaconda3/bin/conda-token
conda-verify: /home/raquellrdc/anaconda3/bin/conda-verify
user site dirs: ~/.local/lib/python3.10
CIO_TEST: <not set>
CONDA_DEFAULT_ENV: kinoml_env
CONDA_EXE: /home/raquellrdc/anaconda3/bin/conda
CONDA_PREFIX: /home/raquellrdc/anaconda3/envs/kinoml_env
CONDA_PROMPT_MODIFIER: (kinoml_env)
CONDA_PYTHON_EXE: /home/raquellrdc/anaconda3/bin/python
CONDA_ROOT: /home/raquellrdc/anaconda3
CONDA_SHLVL: 1
CURL_CA_BUNDLE: <not set>
LD_PRELOAD: <not set>
PATH: /home/raquellrdc/anaconda3/envs/kinoml_env/bin:/home/raquellrdc/anaconda3/condabin:/home/raquellrdc/.local/bin:/usr/local/sbin:/usr/local/bin:/usr/sbin:/usr/bin:/sbin:/bin:/usr/games:/usr/local/games:/snap/bin
PYTHONHASHSEED: 1234
REQUESTS_CA_BUNDLE: <not set>
SSL_CERT_FILE: <not set>
XDG_SEAT_PATH: /org/freedesktop/DisplayManager/Seat0
XDG_SESSION_PATH: /org/freedesktop/DisplayManager/Session1
conda list
----------
# packages in environment at /home/raquellrdc/anaconda3/envs/kinoml_env:
#
# Name Version Build Channel
_libgcc_mutex 0.1 conda_forge conda-forge
_openmp_mutex 4.5 2_gnu conda-forge
_py-xgboost-mutex 2.0 gpu_0 conda-forge
alabaster 0.7.13 pypi_0 pypi
ambertools 23.3 py39h7fa913e_4 conda-forge
amberutils 21.0 pypi_0 pypi
ansiwrap 0.8.4 py_0 conda-forge
anyio 3.5.0 py39h06a4308_0
appdirs 1.4.4 pyh9f0ad1d_0 conda-forge
argon2-cffi 21.3.0 pyhd3eb1b0_0
argon2-cffi-bindings 21.2.0 py39h7f8727e_0
arpack 3.8.0 nompi_h0baa96a_101 conda-forge
arrow 1.3.0 pyhd8ed1ab_0 conda-forge
asttokens 2.0.5 pyhd3eb1b0_0
astunparse 1.6.3 pyhd8ed1ab_0 conda-forge
async-lru 2.0.4 pyhd8ed1ab_0 conda-forge
attrs 23.1.0 py39h06a4308_0
awkward 2.4.6 pyhd8ed1ab_0 conda-forge
awkward-cpp 24 py39h7633fee_0 conda-forge
aws-c-auth 0.7.4 h1083cbe_2 conda-forge
aws-c-cal 0.6.2 h09139f6_2 conda-forge
aws-c-common 0.9.3 hd590300_0 conda-forge
aws-c-compression 0.2.17 h184a658_3 conda-forge
aws-c-event-stream 0.3.2 h6fea174_2 conda-forge
aws-c-http 0.7.13 hb59894b_2 conda-forge
aws-c-io 0.13.33 h161b759_0 conda-forge
aws-c-mqtt 0.9.7 h55cd26b_0 conda-forge
aws-c-s3 0.3.17 hfb4bb88_4 conda-forge
aws-c-sdkutils 0.1.12 h184a658_2 conda-forge
aws-checksums 0.1.17 h184a658_2 conda-forge
aws-crt-cpp 0.24.2 ha28989d_2 conda-forge
aws-sdk-cpp 1.11.156 h314d761_4 conda-forge
babel 2.11.0 py39h06a4308_0
backcall 0.2.0 pyhd3eb1b0_0
beautifulsoup4 4.12.2 py39h06a4308_0
biopandas 0.4.1 pyhd8ed1ab_1 conda-forge
biopython 1.77 py39h27cfd23_0
biotite 0.38.0 py39h44dd56e_0 conda-forge
black 23.3.0 py39h06a4308_0
blas 1.0 mkl
bleach 4.1.0 pyhd3eb1b0_0
blosc 1.21.5 h0f2a231_0 conda-forge
bokeh 3.2.1 py39h2f386ee_0
bravado 11.0.3 pyhd8ed1ab_0 conda-forge
bravado-core 5.17.1 pyhd8ed1ab_0 conda-forge
brotli 1.0.9 he6710b0_2
brotli-python 1.0.9 py39h6a678d5_7
bzip2 1.0.8 h7b6447c_0
c-ares 1.20.1 hd590300_0 conda-forge
c-blosc2 2.10.5 hb4ffafa_0 conda-forge
ca-certificates 2023.08.22 h06a4308_0
cached-property 1.5.2 hd8ed1ab_1 conda-forge
cached_property 1.5.2 pyha770c72_1 conda-forge
cachetools 5.3.1 pyhd8ed1ab_0 conda-forge
cairo 1.18.0 h3faef2a_0 conda-forge
certifi 2023.7.22 py39h06a4308_0
cffi 1.15.1 py39h5eee18b_3
cftime 1.6.2 py39h7deecbd_0
charset-normalizer 2.0.4 pyhd3eb1b0_0
click 8.0.4 py39h06a4308_0
cloudpickle 2.2.1 py39h06a4308_0
codecov 2.1.13 pyhd8ed1ab_0 conda-forge
colorama 0.4.6 py39h06a4308_0
comm 0.1.2 py39h06a4308_0
contourpy 1.0.5 py39hdb19cb5_0
coverage 7.2.2 py39h5eee18b_0
cpuonly 2.0 0 pytorch
cuda-version 11.8 h70ddcb2_2 conda-forge
cudatoolkit 11.8.0 h4ba93d1_12 conda-forge
cycler 0.11.0 pyhd3eb1b0_0
cytoolz 0.12.0 py39h5eee18b_0
dask 2023.10.0 pyhd8ed1ab_0 conda-forge
dask-core 2023.10.0 pyhd8ed1ab_0 conda-forge
dask-jobqueue 0.8.2 pyhd8ed1ab_0 conda-forge
debugpy 1.6.7 py39h6a678d5_0
decorator 5.1.1 pyhd3eb1b0_0
defusedxml 0.7.1 pyhd3eb1b0_0
distributed 2023.10.0 pyhd8ed1ab_0 conda-forge
docker-pycreds 0.4.0 pypi_0 pypi
docutils 0.20.1 pypi_0 pypi
edgembar 0.2 pypi_0 pypi
entrypoints 0.4 py39h06a4308_0
exceptiongroup 1.0.4 py39h06a4308_0
execnet 1.9.0 pyhd3eb1b0_0
executing 0.8.3 pyhd3eb1b0_0
expat 2.5.0 h6a678d5_0
fasteners 0.16.3 pyhd3eb1b0_0
ffmpeg 4.3 hf484d3e_0 pytorch
fftw 3.3.10 nompi_hc118613_108 conda-forge
filelock 3.9.0 py39h06a4308_0
font-ttf-dejavu-sans-mono 2.37 hd3eb1b0_0
font-ttf-inconsolata 2.001 hcb22688_0
font-ttf-source-code-pro 2.030 hd3eb1b0_0
font-ttf-ubuntu 0.83 h8b1ccd4_0
fontconfig 2.14.2 h14ed4e7_0 conda-forge
fonts-anaconda 1 h8fa9717_0
fonts-conda-ecosystem 1 hd3eb1b0_0
fonttools 4.25.0 pyhd3eb1b0_0
fqdn 1.5.1 pyhd8ed1ab_0 conda-forge
freetype 2.12.1 h4a9f257_0
fsspec 2023.9.2 py39h06a4308_0
gettext 0.21.1 h27087fc_0 conda-forge
gflags 2.2.2 he6710b0_0
giflib 5.2.1 h5eee18b_3
gitdb 4.0.11 pypi_0 pypi
gitpython 3.1.40 pypi_0 pypi
glog 0.6.0 h6f12383_0 conda-forge
gmp 6.2.1 h295c915_3
gmpy2 2.1.2 py39heeb90bb_0
gnutls 3.6.15 he1e5248_0
greenlet 2.0.1 py39h6a678d5_0
griddataformats 1.0.1 pyhd8ed1ab_0 conda-forge
gsd 3.2.0 py39h44dd56e_0 conda-forge
h5py 3.10.0 nompi_py39h87cadad_100 conda-forge
hdf4 4.2.15 h9772cbc_5 conda-forge
hdf5 1.14.2 nompi_h4f84152_100 conda-forge
heapdict 1.0.1 pyhd3eb1b0_0
icu 73.2 h59595ed_0 conda-forge
idna 3.4 py39h06a4308_0
imagesize 1.4.1 pypi_0 pypi
importlib-metadata 6.0.0 py39h06a4308_0
importlib-resources 6.1.0 pyhd8ed1ab_0 conda-forge
importlib_metadata 6.0.0 hd3eb1b0_0
importlib_resources 6.1.0 pyhd8ed1ab_0 conda-forge
iniconfig 1.1.1 pyhd3eb1b0_0
intel-openmp 2023.1.0 hdb19cb5_46305
ipykernel 6.25.0 py39h2f386ee_0
ipython 8.15.0 py39h06a4308_0
ipython_genutils 0.2.0 pyhd3eb1b0_1
ipywidgets 8.0.4 py39h06a4308_0
isoduration 20.11.0 pyhd8ed1ab_0 conda-forge
jedi 0.18.1 py39h06a4308_1
jinja2 3.1.2 py39h06a4308_0
joblib 1.2.0 py39h06a4308_0
jpeg 9e h5eee18b_1
json5 0.9.6 pyhd3eb1b0_0
jsonpointer 2.4 py39hf3d152e_3 conda-forge
jsonref 0.2 py_0 conda-forge
jsonschema 4.19.1 pyhd8ed1ab_0 conda-forge
jsonschema-specifications 2023.7.1 pyhd8ed1ab_0 conda-forge
jsonschema-with-format-nongpl 4.19.1 pyhd8ed1ab_0 conda-forge
jupyter-lsp 2.2.0 pyhd8ed1ab_0 conda-forge
jupyter_client 7.4.9 py39h06a4308_0
jupyter_core 5.3.0 py39h06a4308_0
jupyter_events 0.7.0 pyhd8ed1ab_2 conda-forge
jupyter_server 2.7.3 pyhd8ed1ab_1 conda-forge
jupyter_server_terminals 0.4.4 pyhd8ed1ab_1 conda-forge
jupyterlab 4.0.7 pyhd8ed1ab_0 conda-forge
jupyterlab_pygments 0.1.2 py_0
jupyterlab_server 2.25.0 pyhd8ed1ab_0 conda-forge
jupyterlab_widgets 3.0.9 pyhd8ed1ab_0 conda-forge
kinoml 0+unknown pypi_0 pypi
kiwisolver 1.4.4 py39h6a678d5_0
krb5 1.20.1 h143b758_1
lame 3.100 h7b6447c_0
lcms2 2.12 h3be6417_0
ld_impl_linux-64 2.38 h1181459_1
lerc 3.0 h295c915_0
libabseil 20230802.1 cxx17_h59595ed_0 conda-forge
libaec 1.1.2 h59595ed_1 conda-forge
libarrow 13.0.0 h4121bdd_9_cpu conda-forge
libblas 3.9.0 1_h86c2bf4_netlib conda-forge
libboost 1.82.0 h109eef0_2
libboost-python 1.82.0 py39hda80f44_6 conda-forge
libbrotlicommon 1.1.0 hd590300_1 conda-forge
libbrotlidec 1.1.0 hd590300_1 conda-forge
libbrotlienc 1.1.0 hd590300_1 conda-forge
libcrc32c 1.1.2 h6a678d5_0
libcurl 8.4.0 h251f7ec_0
libdeflate 1.17 h5eee18b_1
libedit 3.1.20221030 h5eee18b_0
libev 4.33 h7f8727e_1
libevent 2.1.12 hdbd6064_1
libffi 3.4.4 h6a678d5_0
libgcc-ng 13.2.0 h807b86a_2 conda-forge
libgfortran-ng 13.2.0 h69a702a_2 conda-forge
libgfortran5 13.2.0 ha4646dd_2 conda-forge
libglib 2.78.0 hebfc3b9_0 conda-forge
libgomp 13.2.0 h807b86a_2 conda-forge
libgoogle-cloud 2.12.0 h19a6dae_3 conda-forge
libgrpc 1.58.1 he06187c_2 conda-forge
libiconv 1.17 h166bdaf_0 conda-forge
libidn2 2.3.4 h5eee18b_0
libjpeg-turbo 2.0.0 h9bf148f_0 pytorch
liblapack 3.9.0 5_h92ddd45_netlib conda-forge
libnetcdf 4.9.2 nompi_h80fb2b6_112 conda-forge
libnghttp2 1.57.0 h2d74bed_0
libnsl 2.0.0 h5eee18b_0
libnuma 2.0.16 h0b41bf4_1 conda-forge
libpng 1.6.39 h5eee18b_0
libprotobuf 4.24.3 hf27288f_1 conda-forge
libre2-11 2023.06.02 h7a70373_0 conda-forge
libsodium 1.0.18 h7b6447c_0
libsqlite 3.43.2 h2797004_0 conda-forge
libssh2 1.10.0 hdbd6064_2
libstdcxx-ng 13.2.0 h7e041cc_2 conda-forge
libtasn1 4.19.0 h5eee18b_0
libthrift 0.19.0 hb90f79a_1 conda-forge
libtiff 4.5.1 h6a678d5_0
libunistring 0.9.10 h27cfd23_0
libutf8proc 2.8.0 h166bdaf_0 conda-forge
libuuid 2.38.1 h0b41bf4_0 conda-forge
libwebp 1.3.2 h11a3e52_0
libwebp-base 1.3.2 h5eee18b_0
libxcb 1.15 h7f8727e_0
libxgboost 1.7.6 cuda118h4159b1e_5 conda-forge
libxml2 2.11.5 h232c23b_1 conda-forge
libxslt 1.1.37 h0054252_1 conda-forge
libzip 1.10.1 h2629f0a_3 conda-forge
libzlib 1.2.13 hd590300_5 conda-forge
lightning-utilities 0.9.0 py39h06a4308_0
llvm-openmp 14.0.6 h9e868ea_0
locket 1.0.0 py39h06a4308_0
lxml 4.9.3 py39hed45dcc_1 conda-forge
lz4 4.3.2 py39h5eee18b_0
lz4-c 1.9.4 h6a678d5_0
lzo 2.10 h516909a_1000 conda-forge
markupsafe 2.1.1 py39h7f8727e_0
matplotlib-base 3.8.0 py39he9076e7_2 conda-forge
matplotlib-inline 0.1.6 py39h06a4308_0
mda-xdrlib 0.2.0 pyhd8ed1ab_0 conda-forge
mdanalysis 2.3.0 py39h4661b88_1 conda-forge
mdtraj 1.9.9 py39h031bd0f_0 conda-forge
mistune 0.8.4 py39h27cfd23_1000
mkl 2023.1.0 h213fc3f_46343
mkl-service 2.4.0 py39h5eee18b_1
mkl_fft 1.3.8 py39h5eee18b_0
mkl_random 1.2.4 py39hdb19cb5_0
mmpbsa-py 16.0 pypi_0 pypi
mmtf-python 1.1.3 pyhd8ed1ab_0 conda-forge
monotonic 1.5 py_0
mpc 1.1.0 h10f8cd9_1
mpfr 4.0.2 hb69a4c5_1
mpmath 1.3.0 py39h06a4308_0
mrcfile 1.4.3 pyhd8ed1ab_0 conda-forge
msgpack-python 1.0.3 py39hd09550d_0
munkres 1.1.4 py_0
mypy_extensions 1.0.0 py39h06a4308_0
nbclassic 0.5.5 py39h06a4308_0
nbclient 0.5.13 py39h06a4308_0
nbconvert 6.5.3 pyhd8ed1ab_0 conda-forge
nbconvert-core 6.5.3 pyhd8ed1ab_0 conda-forge
nbconvert-pandoc 6.5.3 pyhd8ed1ab_0 conda-forge
nbformat 5.9.2 py39h06a4308_0
nbval 0.10.0 pyhd8ed1ab_0 conda-forge
nccl 2.19.3.1 h6103f9b_0 conda-forge
ncurses 6.4 h6a678d5_0
nest-asyncio 1.5.6 py39h06a4308_0
netcdf-fortran 4.6.1 nompi_hacb5139_102 conda-forge
netcdf4 1.6.4 nompi_py39h4282601_103 conda-forge
nettle 3.7.3 hbbd107a_1
networkx 3.1 py39h06a4308_0
nglview 3.0.8 pyh1da8cd4_0 conda-forge
notebook 7.0.5 pyhd8ed1ab_0 conda-forge
notebook-shim 0.2.2 py39h06a4308_0
numexpr 2.7.3 py39hde0f152_1 conda-forge
numpy 1.26.0 py39h5f9d8c6_0
numpy-base 1.26.0 py39hb5e798b_0
ocl-icd 2.3.1 h7f98852_0 conda-forge
ocl-icd-system 1.0.0 1 conda-forge
opencadd 0.2.2 pyhd8ed1ab_0 conda-forge
openeye-toolkits 2023.1.1 py39_0 openeye
openff-amber-ff-ports 0.0.3 pyh6c4a22f_0 conda-forge
openff-forcefields 2023.08.0 pyh1a96a4e_0 conda-forge
openff-interchange 0.2.2 pyhd8ed1ab_0 conda-forge
openff-interchange-base 0.2.2 pyhd8ed1ab_0 conda-forge
openff-toolkit 0.11.2 pyhd8ed1ab_2 conda-forge
openff-toolkit-base 0.11.2 pyhd8ed1ab_2 conda-forge
openff-units 0.1.8 pyh1a96a4e_1 conda-forge
openff-utilities 0.1.10 pyhd8ed1ab_0 conda-forge
openh264 2.1.1 h4ff587b_0
openjpeg 2.4.0 h3ad879b_0
openmm 8.0.0 py39he81762f_3 conda-forge
openssl 3.1.3 hd590300_0 conda-forge
orc 1.9.0 h208142c_3 conda-forge
overrides 7.4.0 pyhd8ed1ab_0 conda-forge
packaging 23.1 py39h06a4308_0
packmol 20.010 h86c2bf4_0 conda-forge
packmol-memgen 2023.2.24 pypi_0 pypi
pandas 2.1.1 py39hddac248_1 conda-forge
pandoc 2.19.2 h32600fe_2 conda-forge
pandocfilters 1.5.0 pyhd3eb1b0_0
panedr 0.7.2 pyhd8ed1ab_0 conda-forge
papermill 2.2.2 pyhd8ed1ab_0 conda-forge
parmed 4.1.0 py39h227be39_0 conda-forge
parso 0.8.3 pyhd3eb1b0_0
partd 1.4.0 py39h06a4308_0
pathspec 0.10.3 py39h06a4308_0
pathtools 0.1.2 pypi_0 pypi
pbr 5.11.1 pyhd8ed1ab_0 conda-forge
pcre2 10.40 hc3806b6_0 conda-forge
pdb4amber 22.0 pypi_0 pypi
perl 5.32.1 4_hd590300_perl5 conda-forge
pexpect 4.8.0 pyhd3eb1b0_3
pickleshare 0.7.5 pyhd3eb1b0_1003
pillow 10.0.1 py39ha6cbd5a_0
pint 0.19.2 pyhd8ed1ab_0 conda-forge
pip 23.3 pyhd8ed1ab_0 conda-forge
pixman 0.42.2 h59595ed_0 conda-forge
pkgutil-resolve-name 1.3.10 pyhd8ed1ab_1 conda-forge
platformdirs 3.10.0 py39h06a4308_0
pluggy 1.0.0 py39h06a4308_1
prometheus_client 0.14.1 py39h06a4308_0
prompt-toolkit 3.0.36 py39h06a4308_0
protobuf 4.24.4 pypi_0 pypi
psutil 5.9.0 py39h5eee18b_0
ptyprocess 0.7.0 pyhd3eb1b0_2
pure_eval 0.2.2 pyhd3eb1b0_0
py-cpuinfo 9.0.0 pyhd8ed1ab_0 conda-forge
py-xgboost 1.7.6 cuda118py39h6e70402_5 conda-forge
pyarrow 13.0.0 py39h6925388_9_cpu conda-forge
pycairo 1.21.0 py39h287db57_0
pycparser 2.21 pyhd3eb1b0_0
pydantic 1.10.13 py39hd1e30aa_0 conda-forge
pyedr 0.7.2 pyhd8ed1ab_0 conda-forge
pyg 2.4.0 py39_torch_2.1.0_cpu pyg
pygments 2.15.1 py39h06a4308_1
pymsmt 22.0 pypi_0 pypi
pyparsing 3.0.9 py39h06a4308_0
pyrsistent 0.18.0 py39heee7806_0
pysocks 1.7.1 py39h06a4308_0
pytables 3.9.1 py39hfbd31a7_0 conda-forge
pytest 7.4.2 pyhd8ed1ab_0 conda-forge
pytest-cov 4.1.0 pyhd8ed1ab_0 conda-forge
pytest-xdist 3.3.1 pyhd8ed1ab_0 conda-forge
python 3.9.18 h0755675_0_cpython conda-forge
python-constraint 1.4.0 py_0 conda-forge
python-dateutil 2.8.2 pyhd3eb1b0_0
python-fastjsonschema 2.16.2 py39h06a4308_0
python-json-logger 2.0.7 pyhd8ed1ab_0 conda-forge
python-lmdb 1.4.1 py39h6a678d5_0
python-tzdata 2023.3 pyhd3eb1b0_0
python_abi 3.9 4_cp39 conda-forge
pytorch 2.1.0 py3.9_cpu_0 pytorch
pytorch-lightning 2.1.0 pyhd8ed1ab_0 conda-forge
pytorch-mutex 1.0 cpu pytorch
pytraj 2.0.6 pypi_0 pypi
pytz 2023.3.post1 py39h06a4308_0
pyyaml 6.0 py39h5eee18b_1
pyzmq 25.1.1 py39hb257651_1 conda-forge
rdkit 2023.09.1 py39hce5ca95_0 conda-forge
rdma-core 28.9 h59595ed_1 conda-forge
re2 2023.06.02 h2873b5e_0 conda-forge
readline 8.2 h5eee18b_0
referencing 0.30.2 pyhd8ed1ab_0 conda-forge
reportlab 3.5.67 py39hfdd840d_1
requests 2.31.0 pyhd8ed1ab_0 conda-forge
rfc3339-validator 0.1.4 pyhd8ed1ab_0 conda-forge
rfc3986-validator 0.1.1 pyh9f0ad1d_0 conda-forge
rpds-py 0.10.6 py39h9fdd4d6_0 conda-forge
ruamel.yaml 0.17.35 py39hd1e30aa_0 conda-forge
ruamel.yaml.clib 0.2.6 py39h5eee18b_1
s2n 1.3.54 h06160fa_0 conda-forge
sander 22.0 pypi_0 pypi
scikit-learn 1.3.0 py39h1128e8f_0
scipy 1.11.3 py39h5f9d8c6_0
seaborn 0.12.2 py39h06a4308_0
send2trash 1.8.2 pyh41d4057_0 conda-forge
sentry-sdk 1.32.0 pypi_0 pypi
setproctitle 1.3.3 pypi_0 pypi
setuptools 68.0.0 py39h06a4308_0
simplejson 3.17.6 py39h7f8727e_0
six 1.16.0 pyhd3eb1b0_1
sklearn-pytorch 0.1.0 pypi_0 pypi
smirnoff99frosst 1.1.0 pyh44b312d_0 conda-forge
smmap 5.0.1 pypi_0 pypi
snappy 1.1.10 h9fff704_0 conda-forge
sniffio 1.2.0 py39h06a4308_1
snowballstemmer 2.2.0 pypi_0 pypi
sortedcontainers 2.4.0 pyhd3eb1b0_0
soupsieve 2.5 py39h06a4308_0
sphinx 7.2.6 pypi_0 pypi
sphinxcontrib-applehelp 1.0.7 pypi_0 pypi
sphinxcontrib-devhelp 1.0.5 pypi_0 pypi
sphinxcontrib-htmlhelp 2.0.4 pypi_0 pypi
sphinxcontrib-jsmath 1.0.1 pypi_0 pypi
sphinxcontrib-qthelp 1.0.6 pypi_0 pypi
sphinxcontrib-serializinghtml 1.1.9 pypi_0 pypi
sqlalchemy 2.0.21 py39h5eee18b_0
stack_data 0.2.0 pyhd3eb1b0_0
swagger-spec-validator 3.0.3 pyhd8ed1ab_0 conda-forge
sympy 1.11.1 py39h06a4308_0
tbb 2021.8.0 hdb19cb5_0
tblib 1.7.0 pyhd3eb1b0_0
tenacity 8.2.2 py39h06a4308_0
terminado 0.17.1 py39h06a4308_0
textwrap3 0.9.2 py_0 conda-forge
threadpoolctl 2.2.0 pyh0d69192_0
tidynamics 1.1.2 pyhd8ed1ab_0 conda-forge
tinycss2 1.2.1 py39h06a4308_0
tk 8.6.13 h2797004_0 conda-forge
toml 0.10.2 pyhd3eb1b0_0
tomli 2.0.1 py39h06a4308_0
toolz 0.12.0 py39h06a4308_0
torchaudio 2.1.0 py39_cpu pytorch
torchmetrics 0.11.4 py39h2f386ee_1
torchvision 0.16.0 pypi_0 pypi
tornado 6.3.3 py39h5eee18b_0
tqdm 4.66.1 pyhd8ed1ab_0 conda-forge
traitlets 5.7.1 py39h06a4308_0
types-python-dateutil 2.8.19.14 pyhd8ed1ab_0 conda-forge
typing 3.10.0.0 py39h06a4308_0
typing-extensions 4.7.1 py39h06a4308_0
typing_extensions 4.7.1 py39h06a4308_0
typing_utils 0.1.0 pyhd8ed1ab_0 conda-forge
tzdata 2023c h04d1e81_0
ucx 1.15.0 h64cca9d_0 conda-forge
uri-template 1.3.0 pyhd8ed1ab_0 conda-forge
urllib3 2.0.3 py39h06a4308_0
wandb 0.15.12 pypi_0 pypi
watermark 2.4.3 pyhd8ed1ab_0 conda-forge
wcwidth 0.2.5 pyhd3eb1b0_0
webcolors 1.13 pyhd8ed1ab_0 conda-forge
webencodings 0.5.1 py39h06a4308_1
websocket-client 0.58.0 py39h06a4308_4
wheel 0.41.2 py39h06a4308_0
widgetsnbextension 4.0.5 py39h06a4308_0
xgboost 1.7.6 cuda118py39h6e70402_5 conda-forge
xmltodict 0.13.0 pyhd8ed1ab_0 conda-forge
xorg-kbproto 1.0.7 h7f98852_1002 conda-forge
xorg-libice 1.1.1 hd590300_0 conda-forge
xorg-libsm 1.2.4 h7391055_0 conda-forge
xorg-libx11 1.8.7 h8ee46fc_0 conda-forge
xorg-libxext 1.3.4 h0b41bf4_2 conda-forge
xorg-libxrender 0.9.11 hd590300_0 conda-forge
xorg-libxt 1.3.0 hd590300_1 conda-forge
xorg-renderproto 0.11.1 h7f98852_1002 conda-forge
xorg-xextproto 7.3.0 h0b41bf4_1003 conda-forge
xorg-xproto 7.0.31 h27cfd23_1007
xyzservices 2022.9.0 py39h06a4308_1
xz 5.4.2 h5eee18b_0
yaml 0.2.5 h7b6447c_0
zeromq 4.3.4 h2531618_0
zict 3.0.0 py39h06a4308_0
zipp 3.11.0 py39h06a4308_0
zlib 1.2.13 hd590300_5 conda-forge
zlib-ng 2.0.7 h0b41bf4_0 conda-forge
zstd 1.5.5 hc292b87_0
[38]:
%%capture cap --no-stderr
w = watermark()
[39]:
import json
with open(OUT / "performance.json", "w") as f:
json.dump(metrics, f, default=str, indent=2)
with open(OUT/ "watermark.txt", "w") as f:
f.write(cap.stdout)
with open(OUT / "hparams.json", "w") as f:
json.dump(_hparams, f, default=str, indent=2)
And this is it! We have run an experiment end to end, from obtaining the data to evaluating and saving the model!